1RA6
 
 | |
1RA7
 
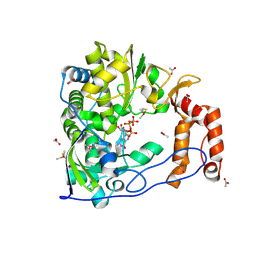 | | Poliovirus Polymerase with GTP | | Descriptor: | ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein | | Authors: | Thompson, A.A, Peersen, O.B. | | Deposit date: | 2003-10-31 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase.
Embo J., 23, 2004
|
|
1RA8
 
 | |
1RA9
 
 | |
1RAJ
 
 | |
1RAK
 
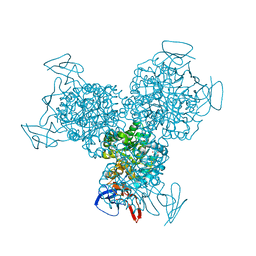 | | Bacterial cytosine deaminase D314S mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RAM
 
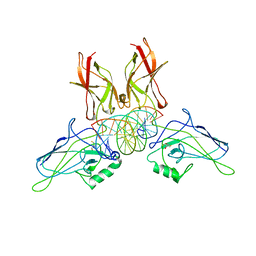 | | A NOVEL DNA RECOGNITION MODE BY NF-KB P65 HOMODIMER | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*CP*GP*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR NF-KB P65) | | Authors: | Chen, Y.-Q, Ghosh, S, Ghosh, G. | | Deposit date: | 1997-11-22 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel DNA recognition mode by the NF-kappa B p65 homodimer.
Nat.Struct.Biol., 5, 1998
|
|
1RAO
 
 | |
1RAP
 
 | |
1RAQ
 
 | |
1RAV
 
 | | RECOMBINANT AVIDIN | | Descriptor: | AVIDIN | | Authors: | Rosano, C, Arosio, P, Bolognesi, M. | | Deposit date: | 1998-03-27 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical characterization and crystal structure of a recombinant hen avidin and its acidic mutant expressed in Escherichia coli.
Eur.J.Biochem., 256, 1998
|
|
1RAY
 
 | |
1RAZ
 
 | |
1RB0
 
 | |
1RB2
 
 | |
1RB3
 
 | |
1RB4
 
 | |
1RB5
 
 | |
1RB6
 
 | |
1RB7
 
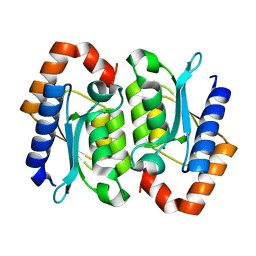 | |
1RB8
 
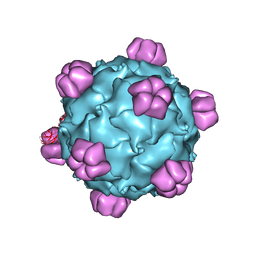 | | The phiX174 DNA binding protein J in two different capsid environments. | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Capsid protein, DNA (5'-D(P*CP*AP*AP*A)-3'), ... | | Authors: | Bernal, R.A, Hafenstein, S, Esmeralda, R, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2003-11-03 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The phiX174 Protein J Mediates DNA Packaging and Viral Attachment to Host Cells.
J.Mol.Biol., 337, 2004
|
|
1RB9
 
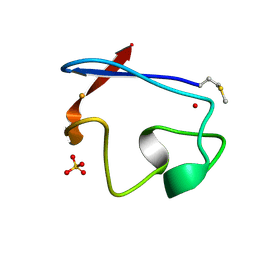 | | RUBREDOXIN FROM DESULFOVIBRIO VULGARIS REFINED ANISOTROPICALLY AT 0.92 ANGSTROMS RESOLUTION | | Descriptor: | FE (II) ION, RUBREDOXIN, SULFATE ION | | Authors: | Dauter, Z, Butterworth, S, Sieker, L.C, Sheldrick, G, Wilson, K.S. | | Deposit date: | 1997-12-21 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Anisotropic Refinement of Rubredoxin from Desulfovibrio Vulgaris
To be Published
|
|
1RBC
 
 | |
1RBD
 
 | |
1RBE
 
 | |
