1J2Z
 
 | | Crystal structure of UDP-N-acetylglucosamine acyltransferase | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Lee, B.I, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Proteins, 53, 2003
|
|
1J30
 
 | | The crystal structure of sulerythrin, a rubrerythrin-like protein from a strictly aerobic and thermoacidiphilic archaeon | | Descriptor: | 144aa long hypothetical rubrerythrin, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-01-16 | | Release date: | 2003-10-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Sulerythrin, A Rubrerythrin-like Protein from A Strictly Aerobic Archaeon, Sulfolobus tokodaii strain 7, shows unexpected domain swapping
Biochemistry, 42, 2003
|
|
1J33
 
 | |
1J34
 
 | | Crystal Structure of Mg(II)-and Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein | | Descriptor: | CALCIUM ION, Coagulation factor IX, MAGNESIUM ION, ... | | Authors: | Shikamoto, Y, Morita, T, Fujimoto, Z, Mizuno, H. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Mg2+- and Ca2+-bound Gla Domain of Factor IX Complexed with Binding Protein
J.Biol.Chem., 278, 2003
|
|
1J35
 
 | | Crystal Structure of Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein | | Descriptor: | CALCIUM ION, Coagulation factor IX, Coagulation factor IX-binding protein B chain, ... | | Authors: | Shikamoto, Y, Morita, T, Fujimoto, Z, Mizuno, H. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Mg2+- and Ca2+-bound Gla Domain of Factor IX Complexed with Binding Protein
J.Biol.Chem., 278, 2003
|
|
1J39
 
 | | Crystal Structure of T4 phage BGT in complex with its UDP-glucose substrate | | Descriptor: | DNA beta-glucosyltransferase, GLYCEROL, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2003-01-21 | | Release date: | 2003-08-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism.
J.Mol.Biol., 330, 2003
|
|
1J3B
 
 | | Crystal structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 | | Descriptor: | ATP-dependent phosphoenolpyruvate carboxykinase, CALCIUM ION, GLYCEROL, ... | | Authors: | Sugahara, M, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1J3E
 
 | | Crystal Structure of the E.coli SeqA protein complexed with N6-methyladenine- guanine mismatch DNA | | Descriptor: | 5'-D(*AP*AP*GP*GP*(6MA)P*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*GP*CP*CP*TP*T)-3', SeqA protein | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-24 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein
Nucleic Acids Res., 32, 2004
|
|
1J3F
 
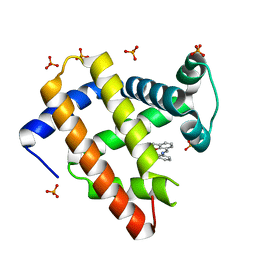 | | Crystal Structure of an Artificial Metalloprotein:Cr(III)(3,3'-Me2-salophen)/apo-A71G Myoglobin | | Descriptor: | 'N,N'-BIS-(2-HYDROXY-3-METHYL-BENZYLIDENE)-BENZENE-1,2-DIAMINE', CHROMIUM ION, Myoglobin, ... | | Authors: | Koshiyama, T, Kono, M, Ohashi, M, Ueno, T, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2003-01-24 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Coordinated Design of Cofactor and Active Site Structures in Development of New Protein Catalysts
J.Am.Chem.Soc., 127, 2005
|
|
1J3H
 
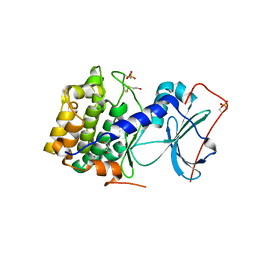 | | Crystal structure of apoenzyme cAMP-dependent protein kinase catalytic subunit | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Wu, J, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic Features of cAMP-dependent Protein Kinase Revealed by Apoenzyme Crystal Structure
J.Mol.Biol., 327, 2003
|
|
1J3I
 
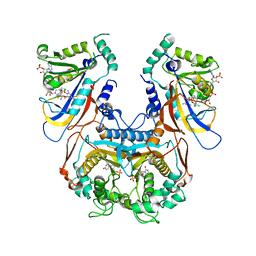 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with WR99210, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 6,6-DIMETHYL-1-[3-(2,4,5-TRICHLOROPHENOXY)PROPOXY]-1,6-DIHYDRO-1,3,5-TRIAZINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuvaniyama, J, Chitnumsub, P, Kamchonwongpaisan, S, Vanichtanankul, J, Sirawaraporn, W, Taylor, P, Walkinshaw, M, Yuthavong, Y. | | Deposit date: | 2003-02-03 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Insights into antifolate resistance from malarial DHFR-TS structures.
NAT.STRUCT.BIOL., 10, 2003
|
|
1J3J
 
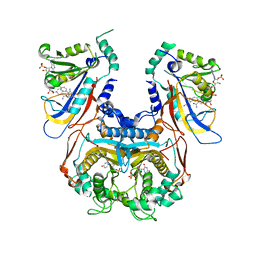 | | Double mutant (C59R+S108N) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with pyrimethamine, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuvaniyama, J, Chitnumsub, P, Kamchonwongpaisan, S, Vanichtanankul, J, Sirawaraporn, W, Taylor, P, Walkinshaw, M, Yuthavong, Y. | | Deposit date: | 2003-02-03 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into antifolate resistance from malarial DHFR-TS structures.
NAT.STRUCT.BIOL., 10, 2003
|
|
1J3K
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with WR99210, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 6,6-DIMETHYL-1-[3-(2,4,5-TRICHLOROPHENOXY)PROPOXY]-1,6-DIHYDRO-1,3,5-TRIAZINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuvaniyama, J, Chitnumsub, P, Kamchonwongpaisan, S, Vanichtanankul, J, Sirawaraporn, W, Taylor, P, Walkinshaw, M, Yuthavong, Y. | | Deposit date: | 2003-02-03 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into antifolate resistance from malarial DHFR-TS structures.
NAT.STRUCT.BIOL., 10, 2003
|
|
1J3L
 
 | | Structure of the RNA-processing inhibitor RraA from Thermus thermophilis | | Descriptor: | CHLORIDE ION, Demethylmenaquinone Methyltransferase, MAGNESIUM ION | | Authors: | Rehse, P.H, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-04 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the RNA-processing inhibitor RraA from Thermus thermophilis.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J3N
 
 | | Crystal Structure of 3-oxoacyl-(acyl-carrier protein) Synthase II from Thermus thermophilus HB8 | | Descriptor: | 3-oxoacyl-(acyl-carrier protein) synthase II, CITRIC ACID, MAGNESIUM ION | | Authors: | Bagautdinov, B, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-10 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-oxoacyl-(acyl-carrier protein) synthase II from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1J3P
 
 | | Crystal structure of Thermococcus litoralis phosphoglucose isomerase | | Descriptor: | FE (III) ION, Phosphoglucose Isomerase | | Authors: | Jeong, J.-J, Fushinobu, S, Ito, S, Hidaka, M, Shoun, H, Wakagi, T. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of a novel cupin-type phosphoglucose isomerase
To be Published
|
|
1J3Q
 
 | | Crystal structure of Thermococcus litoralis phosphogrucose isomerase soaked with FeSO4 | | Descriptor: | FE (III) ION, Phosphoglucose Isomerase | | Authors: | Jeong, J.-J, Fushinobu, S, Ito, S, Hidaka, M, Shoun, H, Wakagi, T. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a novel cupin-type phosphoglucose isomerase
To be Published
|
|
1J3R
 
 | | Crystal structure of Thermococcus litoralis phosphogrucose isomerase complexed with gluconate-6-phosphate | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, FE (III) ION, Phosphoglucose Isomerase | | Authors: | Jeong, J.-J, Fushinobu, S, Ito, S, Hidaka, M, Shoun, H, Wakagi, T. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of a novel cupin-type phosphoglucose isomerase
To be Published
|
|
1J3U
 
 | |
1J3W
 
 | |
1J42
 
 | | Crystal Structure of Human DJ-1 | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Cha, S.S. | | Deposit date: | 2003-02-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain.
J.Biol.Chem., 278, 2003
|
|
1J49
 
 | | INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS | | Descriptor: | D-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Razeto, A, Kochhar, S, Hottinger, H, Dauter, M, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus.
J.Mol.Biol., 318, 2002
|
|
1J4A
 
 | | INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS | | Descriptor: | D-LACTATE DEHYDROGENASE, SULFATE ION | | Authors: | Razeto, A, Kochhar, S, Hottinger, H, Dauter, M, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2001-08-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus.
J.Mol.Biol., 318, 2002
|
|
1J4B
 
 | | Recombinant Mouse-Muscle Adenylosuccinate Synthetase | | Descriptor: | adenylosuccinate synthetase | | Authors: | Iancu, C.V, Borza, T, Choe, J.Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-09-07 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recombinant mouse muscle adenylosuccinate synthetase: overexpression, kinetics, and crystal structure.
J.Biol.Chem., 276, 2001
|
|
1J4E
 
 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE COVALENTLY BOUND TO THE SUBSTRATE DIHYDROXYACETONE PHOSPHATE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE A | | Authors: | Choi, K.H, Shi, J, Hopkins, C.E, Tolan, D.R, Allen, K.N. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate.
Biochemistry, 40, 2001
|
|
