1AEV
 
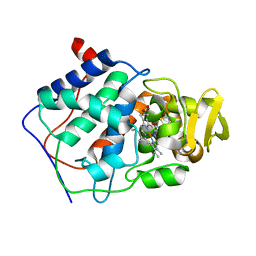 | | INTRODUCTION OF NOVEL SUBSTRATE OXIDATION INTO CYTOCHROME C PEROXIDASE BY CAVITY COMPLEMENTATION: OXIDATION OF 2-AMINOTHIAZOLE AND COVALENT MODIFICATION OF THE ENZYME (2-AMINOTHIAZOLE) | | Descriptor: | 2-AMINOTHIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Fitzgerald, M.M, Jensen, G.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Introduction of novel substrate oxidation into cytochrome c peroxidase by cavity complementation: oxidation of 2-aminothiazole and covalent modification of the enzyme.
Biochemistry, 36, 1997
|
|
1AEW
 
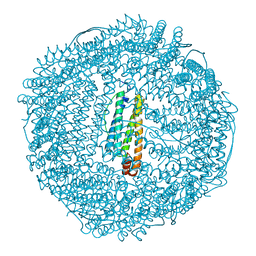 | | L-CHAIN HORSE APOFERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Hempstead, P.D, Yewdall, S.J, Lawson, D.M, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution.
J.Mol.Biol., 268, 1997
|
|
1AF2
 
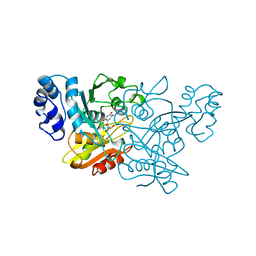 | | CRYSTAL STRUCTURE OF CYTIDINE DEAMINASE COMPLEXED WITH URIDINE | | Descriptor: | CYTIDINE DEAMINASE, URIDINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Xiang, S, Carter, C.W. | | Deposit date: | 1997-03-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the cytidine deaminase-product complex provides evidence for efficient proton transfer and ground-state destabilization.
Biochemistry, 36, 1997
|
|
1AF3
 
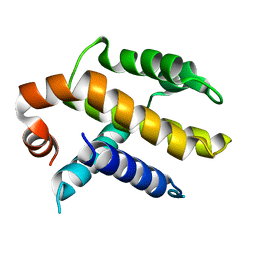 | | RAT BCL-XL AN APOPTOSIS INHIBITORY PROTEIN | | Descriptor: | APOPTOSIS REGULATOR BCL-X | | Authors: | Aritomi, M, Kunishima, N, Inohara, N, Ishibashi, Y, Ohta, S, Morikawa, K. | | Deposit date: | 1997-03-21 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat Bcl-xL. Implications for the function of the Bcl-2 protein family.
J.Biol.Chem., 272, 1997
|
|
1AF4
 
 | | CRYSTAL STRUCTURE OF SUBTILISIN CARLSBERG IN ANHYDROUS DIOXANE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, SUBTILISIN CARLSBERG | | Authors: | Schmitke, J.L, Stern, L.J, Klibanov, A.M. | | Deposit date: | 1997-03-21 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of subtilisin Carlsberg in anhydrous dioxane and its comparison with those in water and acetonitrile.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AF6
 
 | | MALTOPORIN SUCROSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1997-03-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin.
J.Mol.Biol., 272, 1997
|
|
1AF7
 
 | | CHER FROM SALMONELLA TYPHIMURIUM | | Descriptor: | CHEMOTAXIS RECEPTOR METHYLTRANSFERASE CHER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Djordjevic, S, Stock, A.M. | | Deposit date: | 1997-03-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine.
Structure, 5, 1997
|
|
1AF9
 
 | | TETANUS NEUROTOXIN C FRAGMENT | | Descriptor: | TETANUS NEUROTOXIN | | Authors: | Umland, T.C, Wingert, L, Swaminathan, S, Furey, W.F, Schmidt, J.J, Sax, M. | | Deposit date: | 1997-03-24 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the receptor binding fragment HC of tetanus neurotoxin.
Nat.Struct.Biol., 4, 1997
|
|
1AFA
 
 | |
1AFB
 
 | |
1AFD
 
 | |
1AFE
 
 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
1AFQ
 
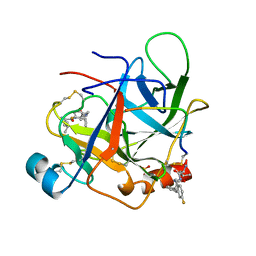 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN COMPLEXED WITH A SYNTHETIC INHIBITOR | | Descriptor: | BOVINE GAMMA-CHYMOTRYPSIN, D-leucyl-N-(4-fluorobenzyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-03-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
1AFV
 
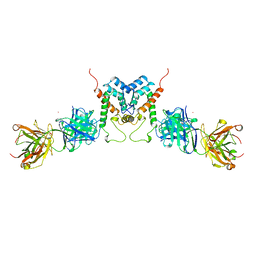 | | HIV-1 CAPSID PROTEIN (P24) COMPLEX WITH FAB25.3 | | Descriptor: | ANTIBODY FAB25.3 FRAGMENT (HEAVY CHAIN), ANTIBODY FAB25.3 FRAGMENT (LIGHT CHAIN), HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 CAPSID PROTEIN, ... | | Authors: | Momany, C, Kovari, L.C, Prongay, A.J, Keller, W, Gitti, R.K, Lee, B.M, Gorbalenya, A.E, Tong, L, Mcclure, J, Ehrlich, L.S, Summers, M.F, Carter, C, Rossmann, M.G. | | Deposit date: | 1997-03-14 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of dimeric HIV-1 capsid protein.
Nat.Struct.Biol., 3, 1996
|
|
1AFW
 
 | |
1AG8
 
 | |
1AG9
 
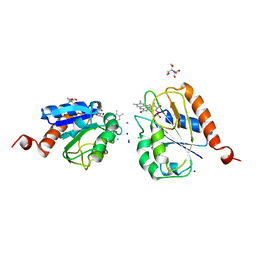 | |
1AGN
 
 | | X-RAY STRUCTURE OF HUMAN SIGMA ALCOHOL DEHYDROGENASE | | Descriptor: | ACETATE ION, HUMAN SIGMA ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hurley, T.D, Xie, P. | | Deposit date: | 1996-06-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of human class IV sigmasigma alcohol dehydrogenase. Structural basis for substrate specificity.
J.Biol.Chem., 272, 1997
|
|
1AGX
 
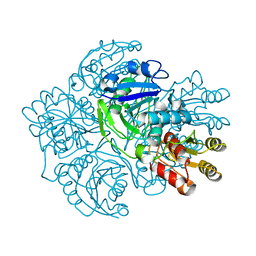 | | REFINED CRYSTAL STRUCTURE OF ACINETOBACTER GLUTAMINASIFICANS GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Housset, D, Weber, I.T, Ammon, H.L, Murphy, K.C, Swain, A.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of Acinetobacter glutaminasificans glutaminase-asparaginase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1AGY
 
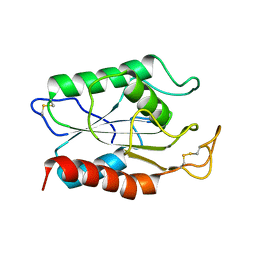 | |
1AH0
 
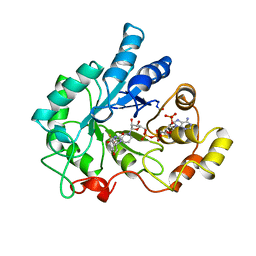 | | PIG ALDOSE REDUCTASE COMPLEXED WITH SORBINIL | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SORBINIL | | Authors: | Moras, D, Podjarny, A.D. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
Structure, 5, 1997
|
|
1AH3
 
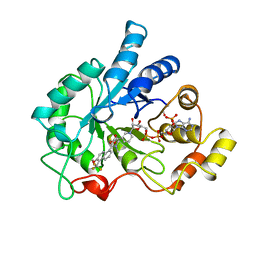 | | ALDOSE REDUCTASE COMPLEXED WITH TOLRESTAT INHIBITOR | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Moras, D, Podjarny, A. | | Deposit date: | 1997-04-12 | | Release date: | 1998-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
Structure, 5, 1997
|
|
1AH4
 
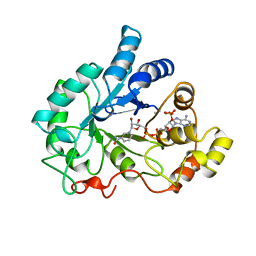 | | PIG ALDOSE REDUCTASE, HOLO FORM | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Moras, D, Podjarny, A. | | Deposit date: | 1997-04-12 | | Release date: | 1998-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
Structure, 5, 1997
|
|
1AH5
 
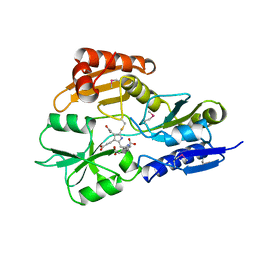 | | REDUCED FORM SELENOMETHIONINE-LABELLED HYDROXYMETHYLBILANE SYNTHASE DETERMINED BY MAD | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Harrop, S.J, Cassetta, A. | | Deposit date: | 1997-04-13 | | Release date: | 1997-10-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AHJ
 
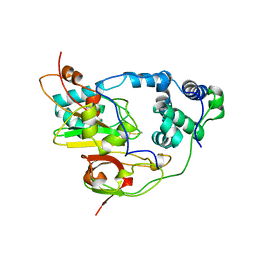 | | NITRILE HYDRATASE | | Descriptor: | FE (III) ION, NITRILE HYDRATASE (SUBUNIT ALPHA), NITRILE HYDRATASE (SUBUNIT BETA) | | Authors: | Huang, W, Schneider, G, Lindqvist, Y. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold.
Structure, 5, 1997
|
|
