1EQM
 
 | | CRYSTAL STRUCTURE OF BINARY COMPLEX OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE WITH ADENOSINE-5'-DIPHOSPHATE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiao, B, Blaszczyk, J, Ji, X. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual conformational changes in 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as revealed by X-ray crystallography and NMR.
J.Biol.Chem., 276, 2001
|
|
1EQN
 
 | | E.COLI PRIMASE CATALYTIC CORE | | Descriptor: | DNA PRIMASE | | Authors: | Podobnik, M, McInerney, P, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2000-04-05 | | Release date: | 2000-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases.
J.Mol.Biol., 300, 2000
|
|
1EQP
 
 | |
1EQQ
 
 | | SINGLE STRANDED DNA BINDING PROTEIN AND SSDNA COMPLEX | | Descriptor: | 5'-R(*(5MU)P*(5MU)P*(5MU))-3', SINGLE STRANDED DNA BINDING PROTEIN | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Yasuoka, N, Shimamoto, N. | | Deposit date: | 2000-04-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis
J.Biochem.(Tokyo), 127, 2000
|
|
1EQR
 
 | | CRYSTAL STRUCTURE OF FREE ASPARTYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTYL-TRNA SYNTHETASE, MAGNESIUM ION | | Authors: | Rees, B, Webster, G, Delarue, M, Boeglin, M, Moras, D. | | Deposit date: | 2000-04-06 | | Release date: | 2000-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aspartyl tRNA-synthetase from Escherichia coli: flexibility and adaptability to the substrates.
J.Mol.Biol., 299, 2000
|
|
1EQT
 
 | | MET-RANTES | | Descriptor: | SULFATE ION, T-CELL SPECIFIC RANTES PROTEIN | | Authors: | Hoover, D.M, Shaw, J, Gryczynski, Z, Proudfoot, A.E.I, Wells, T. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of MET-RANTES: Comparison with Native RANTES and AOP-RANTES
PROTEIN PEPT.LETT., 7, 2000
|
|
1EQV
 
 | |
1EQW
 
 | | CRYSTAL STRUCTURE OF SALMONELLA TYPHIMURIUM CU,ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Pesce, A, Battistoni, A, Stroppolo, M.E, Polizio, F, Nardini, M, Kroll, J.S, Langford, P.R, O'Neill, P, Sette, M, Desideri, A, Bolognesi, M. | | Deposit date: | 2000-04-06 | | Release date: | 2000-09-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and crystallographic characterization of Salmonella typhimurium Cu,Zn superoxide dismutase coded by the sodCI virulence gene.
J.Mol.Biol., 302, 2000
|
|
1ERE
 
 | |
1ERJ
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL WD40 DOMAIN OF TUP1 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR TUP1 | | Authors: | Sprague, E.R, Redd, M.J, Johnson, A.D, Wolberger, C. | | Deposit date: | 2000-04-06 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast.
EMBO J., 19, 2000
|
|
1ERR
 
 | |
1ERZ
 
 | | CRYSTAL STRUCTURE OF N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE WITH A NOVEL CATALYTIC FRAMEWORK COMMON TO AMIDOHYDROLASES | | Descriptor: | N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE | | Authors: | Nakai, T, Hasegawa, T, Yamashita, E, Yamamoto, M, Kumasaka, T, Ueki, T, Nanba, H, Ikenaka, Y, Takahashi, S, Sato, M, Tsukihara, T. | | Deposit date: | 2000-04-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases.
Structure Fold.Des., 8, 2000
|
|
1ES0
 
 | |
1ES1
 
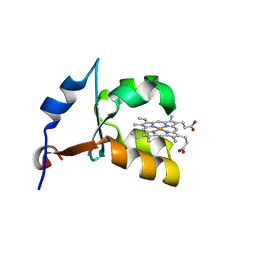 | | CRYSTAL STRUCTURE OF VAL61HIS MUTANT OF TRYPSIN-SOLUBILIZED FRAGMENT OF CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, J, Gan, J.-H, Xia, Z.-X, Wang, Y.-H, Wang, W.-H, Xue, L.-L, Xie, Y, Huang, Z.-X. | | Deposit date: | 2000-04-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
Proteins, 40, 2000
|
|
1ES2
 
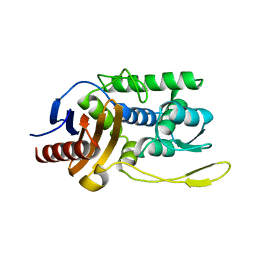 | | S96A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES3
 
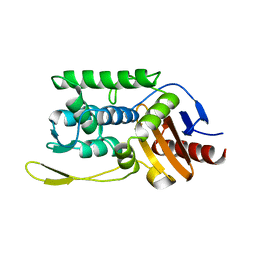 | | C98A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE, SODIUM ION | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES4
 
 | | C98N mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES5
 
 | |
1ES6
 
 | | CRYSTAL STRUCTURE OF THE MATRIX PROTEIN OF EBOLA VIRUS | | Descriptor: | MATRIX PROTEIN VP40 | | Authors: | Dessen, A, Volchkov, V, Dolnik, O, Klenk, H.-D, Weissenhorn, W. | | Deposit date: | 2000-04-07 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the matrix protein VP40 from Ebola virus.
EMBO J., 19, 2000
|
|
1ES9
 
 | | X-RAY CRYSTAL STRUCTURE OF R22K MUTANT OF THE MAMMALIAN BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASES (PAF-AH) | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB GAMMA SUBUNIT | | Authors: | McMullen, T.W.P, Li, J, Sheffield, P.J, Aoki, J, Martin, T.W, Arai, H, Inoue, K, Derewenda, Z.S. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib).
Protein Eng., 13, 2000
|
|
1ESF
 
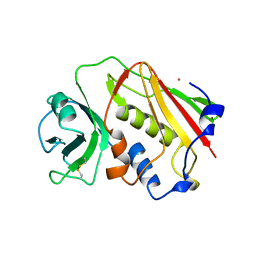 | | STAPHYLOCOCCAL ENTEROTOXIN A | | Descriptor: | CADMIUM ION, STAPHYLOCOCCAL ENTEROTOXIN A | | Authors: | Schad, E.M, Svensson, L.A. | | Deposit date: | 1995-05-25 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superantigen staphylococcal enterotoxin type A.
EMBO J., 14, 1995
|
|
1ESG
 
 | |
1ESI
 
 | |
1ESJ
 
 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) | | Descriptor: | HYDROXYETHYLTHIAZOLE KINASE, SULFATE ION | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
1ESM
 
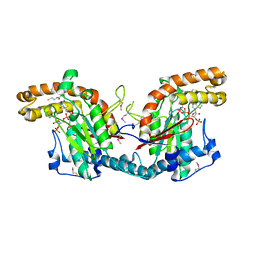 | | STRUCTURAL BASIS FOR THE FEEDBACK REGULATION OF ESCHERICHIA COLI PANTOTHENATE KINASE BY COENZYME A | | Descriptor: | COENZYME A, PANTOTHENATE KINASE | | Authors: | Yun, M, Park, C.G, Kim, J.Y, Rock, C.O, Jackowski, S, Park, H.W. | | Deposit date: | 2000-04-10 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A.
J.Biol.Chem., 275, 2000
|
|
