1DL5
 
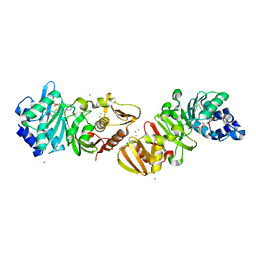 | | PROTEIN-L-ISOASPARTATE O-METHYLTRANSFERASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, PROTEIN-L-ISOASPARTATE O-METHYLTRANSFERASE, ... | | Authors: | Skinner, M.M, Puvathingal, J.M, Walter, R.L, Friedman, A.M. | | Deposit date: | 1999-12-08 | | Release date: | 2000-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of protein isoaspartyl methyltransferase: a catalyst for protein repair.
Structure Fold.Des., 8, 2000
|
|
1DL8
 
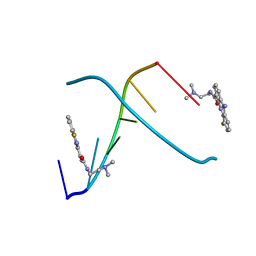 | | CRYSTAL STRUCTURE OF 5-F-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 5-FLUORO-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides.
Mol.Pharmacol., 58, 2000
|
|
1DLA
 
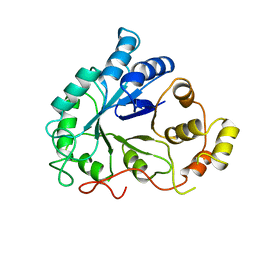 | | NOVEL NADPH-BINDING DOMAIN REVEALED BY THE CRYSTAL STRUCTURE OF ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE | | Authors: | Rondeau, J.-M, Tete-Favier, F, Podjarny, A, Reymann, J.-M, Barth, P, Biellmann, J.-F, Moras, D. | | Deposit date: | 1993-02-08 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel NADPH-binding domain revealed by the crystal structure of aldose reductase.
Nature, 355, 1992
|
|
1DLE
 
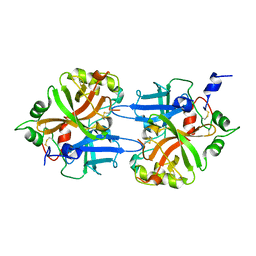 | | FACTOR B SERINE PROTEASE DOMAIN | | Descriptor: | COMPLEMENT FACTOR B | | Authors: | Jing, H, Xu, Y, Carson, M, Moore, D, Macon, K.J, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 1999-12-09 | | Release date: | 2000-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New structural motifs on the chymotrypsin fold and their potential roles in complement factor B.
EMBO J., 19, 2000
|
|
1DLK
 
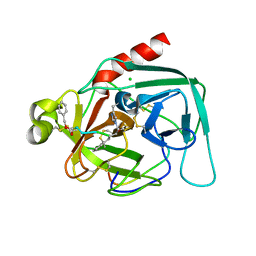 | | CRYSTAL STRUCTURE ANALYSIS OF DELTA-CHYMOTRYPSIN BOUND TO A PEPTIDYL CHLOROMETHYL KETONE INHIBITOR | | Descriptor: | CHLORIDE ION, Thrombin heavy chain, Thrombin light chain, ... | | Authors: | Mac Sweeney, A, Birrane, G, Walsh, M.A, O'Connell, T, Malthouse, J.P.G. | | Deposit date: | 1999-12-10 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of delta-chymotrypsin bound to a peptidyl chloromethyl ketone inhibitor.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DLL
 
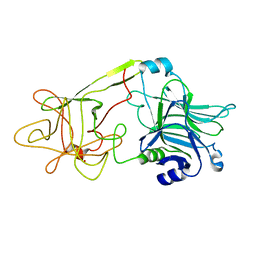 | | The HC fragement of tetanus toxin complexed with lactose | | Descriptor: | GLYCEROL, TETANUS TOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-12-10 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
1DLM
 
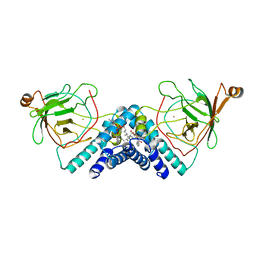 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER CALCOACETICUS NATIVE DATA | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, [1-PENTADECANOYL-2-DECANOYL-GLYCEROL-3-YL]PHOSPHONYL CHOLINE | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DLQ
 
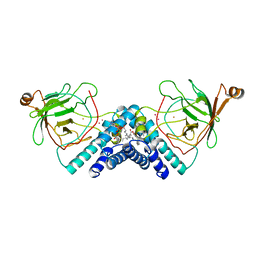 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 INHIBITED BY BOUND MERCURY | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DLT
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND CATECHOL | | Descriptor: | CATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-12 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DLU
 
 | | UNLIGANDED BIOSYNTHETIC THIOLASE FROM ZOOGLOEA RAMIGERA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIOSYNTHETIC THIOLASE, SULFATE ION | | Authors: | Modis, Y, Wierenga, R.K. | | Deposit date: | 1999-12-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase.
J.Mol.Biol., 297, 2000
|
|
1DLV
 
 | |
1DM3
 
 | |
1DM5
 
 | |
1DM6
 
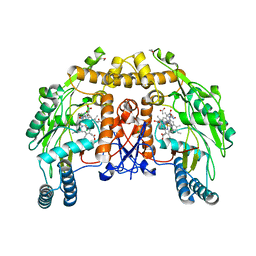 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH N-(4-CHLOROPHENYL)-N'-HYDROXYGUANIDINE (H4B FREE) | | Descriptor: | ACETATE ION, CACODYLATE ION, N-(CHLOROPHENYL)-N'-HYDROXYGUANIDINE, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-12-13 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
1DM7
 
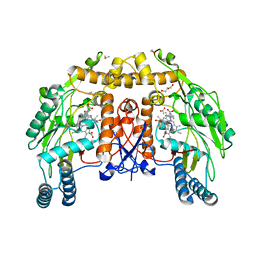 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH HOMOARGININE (H4B FREE) | | Descriptor: | ACETATE ION, CACODYLATE ION, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-12-13 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Bovine Endothelial Nitric Oxide Synthase Heme Domain Complexed With Various Inhibitors
To be Published
|
|
1DM8
 
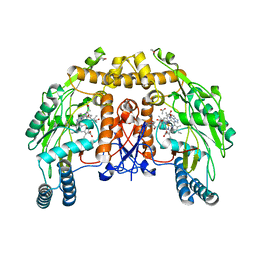 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 1,2,4-TRIAZOLE-CARBOXAMIDINE (H4B BOUND) | | Descriptor: | 1,2,4-TRIAZOLE-CARBOXAMIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-12-13 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of Bovine Endothelial Nitric Oxide Synthase Heme Domain Complexed With Various Inhibitors
To be Published
|
|
1DMB
 
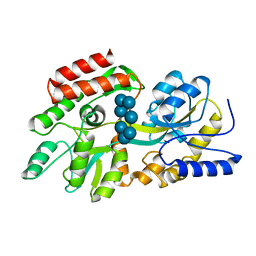 | |
1DMH
 
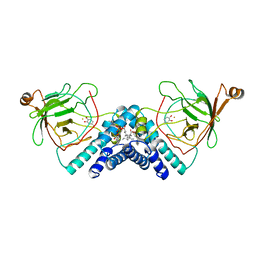 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND 4-METHYLCATECHOL | | Descriptor: | 4-METHYLCATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DMI
 
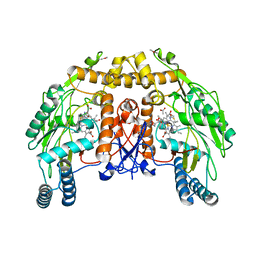 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 6S-H4B | | Descriptor: | 6S-5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kotsonis, P, Pfleiderer, W, Schmidt, H.H.H.W, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Bovine Endothelial Nitric Oxide Synthase Heme Domain Complexed with the Pterin Analogues
To be Published
|
|
1DMJ
 
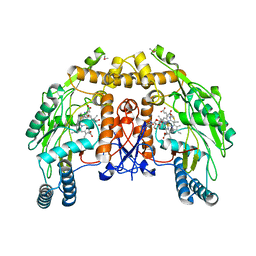 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 5,6-CYCLIC-TETRAHYDROPTERIDINE | | Descriptor: | 7-AMINO-3,3A,4,5-TETRAHYDRO-8H-2-OXA-5,6,8,9B-TETRAAZA-CYCLOPENTA[A]NAPHTHALENE-1,9-DIONE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
1DMK
 
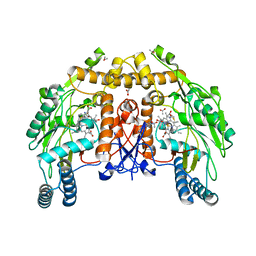 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 4-AMINO-6-PHENYL-TETRAHYDROPTERIDINE | | Descriptor: | 2,4-DIAMINO-6-PHENYL-5,6,7,8,-TETRAHYDROPTERIDINE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
1DML
 
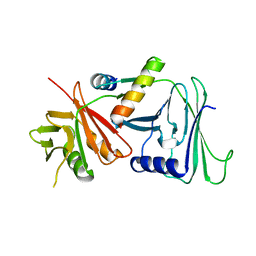 | | CRYSTAL STRUCTURE OF HERPES SIMPLEX UL42 BOUND TO THE C-TERMINUS OF HSV POL | | Descriptor: | DNA POLYMERASE, DNA POLYMERASE PROCESSIVITY FACTOR | | Authors: | Zuccola, H.J, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 1999-12-14 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase.
Mol.Cell, 5, 2000
|
|
1DMM
 
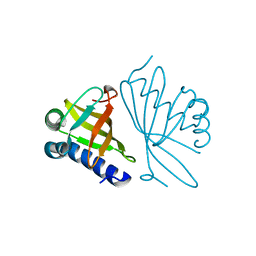 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMN
 
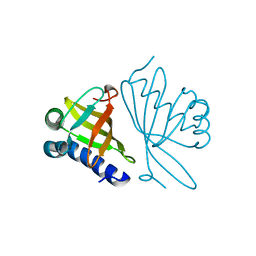 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMQ
 
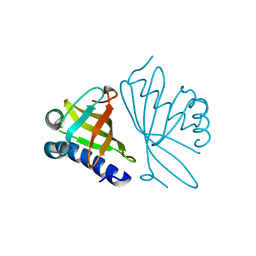 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
