1SMB
 
 | | Crystal Structure of Golgi-Associated PR-1 protein | | Descriptor: | 17kD fetal brain protein | | Authors: | Serrano, R.L, Kuhn, A, Hendricks, A, Helms, J.B, Sinning, I, Groves, M.R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of the human Golgi-associated plant pathogenesis related protein GAPR-1 implicates dimerization as a regulatory mechanism
J.Mol.Biol., 339, 2004
|
|
1SMC
 
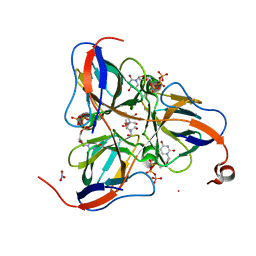 | | Mycobacterium tuberculosis dUTPase complexed with dUTP in the absence of metal ion. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SMD
 
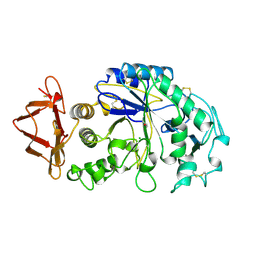 | | HUMAN SALIVARY AMYLASE | | Descriptor: | AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Ramasubbu, N. | | Deposit date: | 1996-01-24 | | Release date: | 1996-07-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1SMF
 
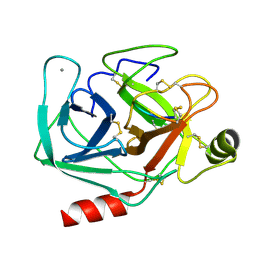 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | Authors: | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | Deposit date: | 1992-10-24 | | Release date: | 1994-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
1SMI
 
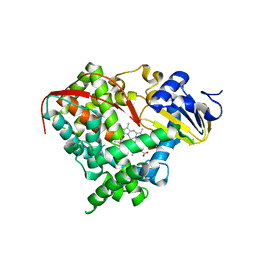 | | A single mutation of P450 BM3 induces the conformational rearrangement seen upon substrate-binding in wild-type enzyme | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Joyce, M.G, Girvan, H.M, Munro, A.W, Leys, D. | | Deposit date: | 2004-03-09 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Single Mutation in Cytochrome P450 BM3 Induces the Conformational Rearrangement Seen upon Substrate Binding in the Wild-type Enzyme
J.Biol.Chem., 279, 2004
|
|
1SMJ
 
 | | Structure of the A264E mutant of cytochrome P450 BM3 complexed with palmitoleate | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Joyce, M.G, Girvan, H.M, Munro, A.W, Leys, D. | | Deposit date: | 2004-03-09 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Single Mutation in Cytochrome P450 BM3 Induces the Conformational Rearrangement Seen upon Substrate Binding in the Wild-type Enzyme
J.Biol.Chem., 279, 2004
|
|
1SMK
 
 | | Mature and translocatable forms of glyoxysomal malate dehydrogenase have different activities and stabilities but similar crystal structures | | Descriptor: | CITRIC ACID, Malate dehydrogenase, glyoxysomal | | Authors: | Cox, B, Chit, M.M, Weaver, T, Bailey, J, Gietl, C, Bell, E, Banaszak, L. | | Deposit date: | 2004-03-09 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence.
Febs J., 272, 2005
|
|
1SML
 
 | | METALLO BETA LACTAMASE L1 FROM STENOTROPHOMONAS MALTOPHILIA | | Descriptor: | PROTEIN (PENICILLINASE), ZINC ION | | Authors: | Ullah, J.H, Walsh, T.R, Taylor, I.A, Emery, D.C, Verma, C.S, Gamblin, S.J, Spencer, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution.
J.Mol.Biol., 284, 1998
|
|
1SMM
 
 | | Crystal Structure of Cp Rd L41A mutant in oxidized state | | Descriptor: | FE (III) ION, Rubredoxin, SULFATE ION | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1SMO
 
 | | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47 . | | Descriptor: | L(+)-TARTARIC ACID, triggering receptor expressed on myeloid cells 1 | | Authors: | Kelker, M.S, Foss, T.R, Peti, W, Teyton, L, Kelly, J.W, Wilson, I.A. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47A.
J.Mol.Biol., 342, 2004
|
|
1SMT
 
 | | SMTB REPRESSOR FROM SYNECHOCOCCUS PCC7942 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR SMTB | | Authors: | Cook, W.J, Hall, L.M. | | Deposit date: | 1997-09-16 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins.
J.Mol.Biol., 275, 1998
|
|
1SMU
 
 | | Crystal Structure of Cp Rd L41A mutant in reduced state 1 (drop-reduced) | | Descriptor: | FE (II) ION, Rubredoxin | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1SMV
 
 | |
1SMW
 
 | | Crystal Structure of Cp Rd L41A mutant in reduced state 2 (soaked) | | Descriptor: | FE (II) ION, Rubredoxin | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1SMX
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SMY
 
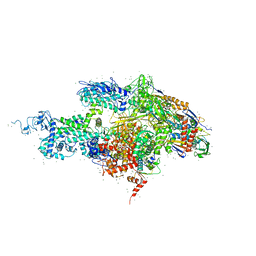 | | Structural basis for transcription regulation by alarmone ppGpp | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
1SN0
 
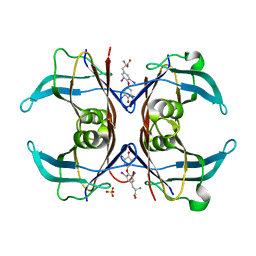 | | Crystal Structure Of Sea Bream Transthyretin in complex with thyroxine At 1.9A Resolution | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Santos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1SN2
 
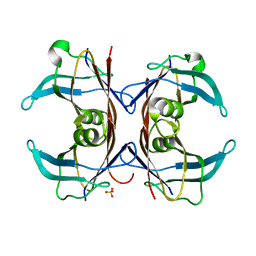 | | Crystal Structure of Sea Bream Transthyretin at 1.90A Resolution | | Descriptor: | SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Cantos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1SN5
 
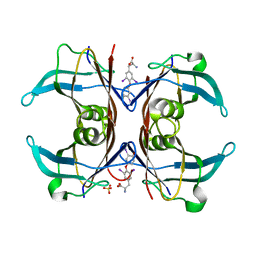 | | Crystal Structure of Sea Bream Transthyretin in complex with Triiodothyronine at 1.90A Resolution | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Cantos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1SN7
 
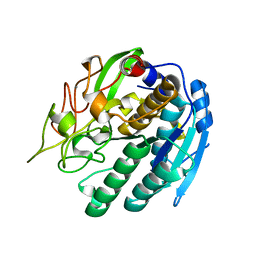 | | KUMAMOLISIN-AS, APOENZYME | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Oda, K, Nishino, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-as, a serine-carboxyl peptidase with collagenase activity.
J.Biol.Chem., 279, 2004
|
|
1SN8
 
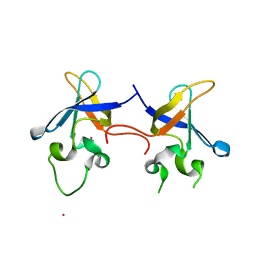 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | Descriptor: | LEAD (II) ION, Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SN9
 
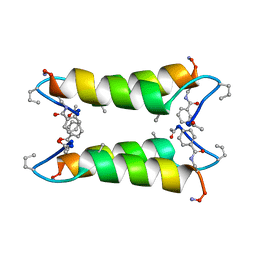 | |
1SNA
 
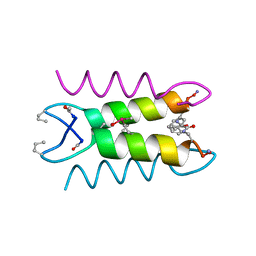 | | An Oligomeric Domain-Swapped Beta-Beta-Alpha Mini-Protein | | Descriptor: | ISOPROPYL ALCOHOL, tetrameric beta-beta-alpha mini-protein | | Authors: | Ali, M.H, Peisach, E, Allen, K.N, Imperiali, B. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SNC
 
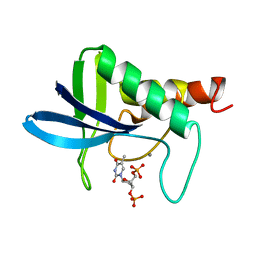 | | THE CRYSTAL STRUCTURE OF THE TERNARY COMPLEX OF STAPHYLOCOCCAL NUCLEASE, CA2+, AND THE INHIBITOR PD*TP, REFINED AT 1.65 ANGSTROMS | | Descriptor: | CALCIUM ION, THERMONUCLEASE PRECURSOR, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Loll, P.J, Lattman, E.E. | | Deposit date: | 1989-07-21 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the ternary complex of staphylococcal nuclease, Ca2+, and the inhibitor pdTp, refined at 1.65 A.
Proteins, 5, 1989
|
|
1SNE
 
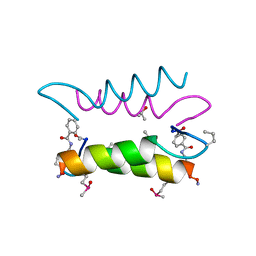 | | An Oligomeric Domain-Swapped Beta-Beta-Alpha Mini-Protein | | Descriptor: | ISOPROPYL ALCOHOL, tetrameric beta-beta-alpha mini-protein | | Authors: | Ali, M.H, Peisach, E, Allen, K.N, Imperiali, B. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
