1P0E
 
 | | CRYSTAL STRUCTURE OF ZYMOMONAS MOBILIS tRNA-GUANINE TRANSGLYCOSYLASE (TGT) COCRYSTALLISED WITH PREQ1 AT PH 5.5 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase
and their implications for substrate selectivity, reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
1P0F
 
 | | Crystal Structure of the Binary Complex: NADP(H)-Dependent Vertebrate Alcohol Dehydrogenase (ADH8) with the cofactor NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent ALCOHOL DEHYDROGENASE, ... | | Authors: | Rosell, A, Valencia, E, Pares, X, Fita, I, Farres, J, Ochoa, W.F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the vertebrate NADP(H)-dependent alcohol dehydrogenase (ADH8)
J.Mol.Biol., 330, 2003
|
|
1P0H
 
 | | Crystal Structure of Rv0819 from Mycobacterium Tuberculosis MshD-Mycothiol Synthase Coenzyme A Complex | | Descriptor: | ACETYL COENZYME *A, COENZYME A, hypothetical protein Rv0819 | | Authors: | Vetting, M.W, Roderick, S.L, Yu, M, Blanchard, J.S. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases.
Protein Sci., 12, 2003
|
|
1P0I
 
 | | Crystal structure of human butyryl cholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
1P0K
 
 | | IPP:DMAPP isomerase type II apo structure | | Descriptor: | Isopentenyl-diphosphate delta-isomerase, PHOSPHATE ION | | Authors: | Steinbacher, S, Kaiser, J, Gerhardt, S, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Type II Isopentenyl Diphosphate:Dimethylallyl Diphosphate Isomerase from Bacillus subtilis
J.Mol.Biol., 329, 2003
|
|
1P0M
 
 | | Crystal structure of human butyryl cholinesterase in complex with a choline molecule | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
1P0N
 
 | | IPP:DMAPP isomerase type II, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Isopentenyl-diphosphate delta-isomerase | | Authors: | Steinbacher, S, Kaiser, J, Gerhardt, S, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Type II Isopentenyl Diphosphate:Dimethylallyl Diphosphate Isomerase from Bacillus subtilis
J.Mol.Biol., 329, 2003
|
|
1P0P
 
 | | Crystal structure of soman-aged human butyryl cholinesterase in complex with the substrate analog butyrylthiocholine | | Descriptor: | 2-(BUTYRYLSULFANYL)-N,N,N-TRIMETHYLETHANAMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
1P0Q
 
 | | Crystal structure of soman-aged human butyryl cholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
1P0S
 
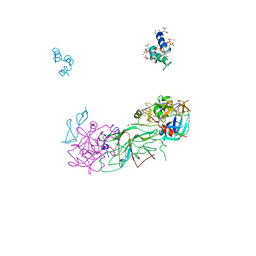 | | Crystal Structure of Blood Coagulation Factor Xa in Complex with Ecotin M84R | | Descriptor: | Coagulation factor X precursor, Ecotin precursor, MAGNESIUM ION, ... | | Authors: | Wang, S.X, Hur, E, Sousa, C.A, Brinen, L, Slivka, E.J, Fletterick, R.J. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Extended Interactions and Gla Domain of Blood Coagulation Factor Xa
Biochemistry, 42, 2003
|
|
1P0Y
 
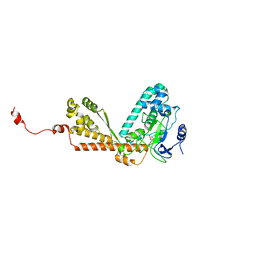 | | Crystal structure of the SET domain of LSMT bound to MeLysine and AdoHcy | | Descriptor: | N-METHYL-LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | Authors: | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
1P0Z
 
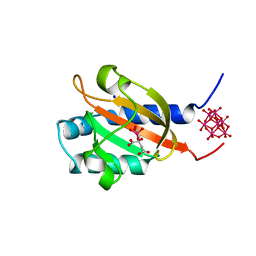 | | Sensor Kinase CitA binding domain | | Descriptor: | CITRATE ANION, MO(VI)(=O)(OH)2 CLUSTER, SODIUM ION, ... | | Authors: | Reinelt, S, Hofmann, E, Gerharz, T, Bott, M, Madden, D.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain.
J.Biol.Chem., 278, 2003
|
|
1P14
 
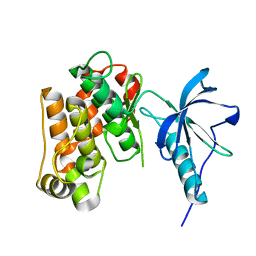 | | Crystal structure of a catalytic-loop mutant of the insulin receptor tyrosine kinase | | Descriptor: | insulin receptor | | Authors: | Li, S, Covino, N.D, Stein, E.G, Till, J.H, Hubbard, S.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical evidence for an autoinhibitory role for tyrosine 984 in the juxtamembrane region of the insulin receptor
J.Biol.Chem., 278, 2003
|
|
1P15
 
 | |
1P16
 
 | | Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Fabrega, C, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II.
Mol.Cell, 11, 2003
|
|
1P1F
 
 | |
1P1H
 
 | | Crystal structure of the 1L-myo-inositol/NAD+ complex | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jin, X, Geiger, J.H. | | Deposit date: | 2003-04-12 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P1I
 
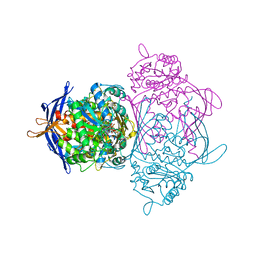 | |
1P1J
 
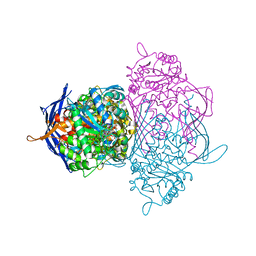 | | Crystal structure of the 1L-myo-inositol 1-phosphate synthase complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Inositol-3-phosphate synthase, ... | | Authors: | Jin, X, Geiger, J.H. | | Deposit date: | 2003-04-12 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P1K
 
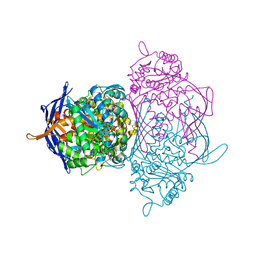 | |
1P1L
 
 | |
1P1M
 
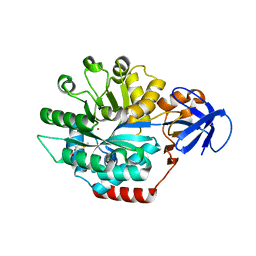 | | Structure of Thermotoga maritima amidohydrolase TM0936 bound to Ni and methionine | | Descriptor: | Hypothetical protein TM0936, METHIONINE, NICKEL (II) ION | | Authors: | Kniewel, R, Buglino, J.A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the hypothetical protein TM0936 from Thermotoga maritima at
1.5A bound to Ni and methionine
To be Published, 2003
|
|
1P1N
 
 | |
1P1O
 
 | | Crystal structure of the GluR2 ligand-binding core (S1S2J) mutant L650T in complex with quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Armstrong, N, Mayer, M.L, Gouaux, E. | | Deposit date: | 2003-04-13 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic
adjustment of agonist-induced conformational changes.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P1Q
 
 | |
