1OYW
 
 | |
1OYX
 
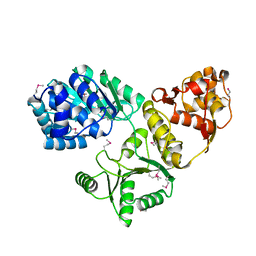
 | | CRYSTAL STRUCTURE OF 3-MBT REPEATS OF LETHAL (3) MALIGNANT BRAIN TUMOR (SELENO-MET) AT 1.85 ANGSTROM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
1OYY
 
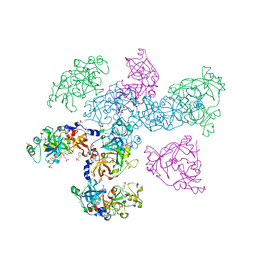
 | | Structure of the RecQ Catalytic Core bound to ATP-gamma-S | | Descriptor: | ATP-dependent DNA helicase, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Bernstein, D.A, Zittel, M.C, Keck, J.L. | | Deposit date: | 2003-04-07 | | Release date: | 2003-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-resolution structure of the E. coli RecQ helicase catalytic core
Embo J., 22, 2003
|
|
1OYZ
 
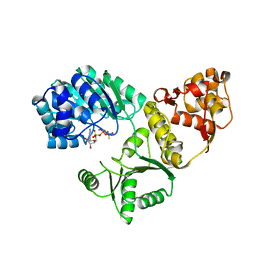
 | | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31. | | Descriptor: | Hypothetical protein yibA | | Authors: | Kuzin, A, Semesi, A, Skarina, T, Savchenko, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31.
To be Published
|
|
1OZ0
 
 | | CRYSTAL STRUCTURE OF THE HOMODIMERIC BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME AVIAN ATIC IN COMPLEX WITH A MULTISUBSTRATE ADDUCT INHIBITOR BETA-DADF. | | Descriptor: | 2-[4-((2-AMINO-4-OXO-3,4-DIHYDRO-PYRIDO[3,2-D]PYRIMIDIN-6-YLMETHYL)-{3-[5-CARBAMOYL-3-(3,4- DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-3H-IMIDAZOL-4-YL]-ACRYLOYL}-AMINO)-BENZOYLAMINO]- PENTANEDIOIC ACID, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Wolan, D.W, Greasley, S.E, Wall, M.J, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Avian AICAR Transformylase with a Multisubstrate Adduct Inhibitor beta-DADF Identifies the Folate Binding Site.
Biochemistry, 42, 2003
|
|
1OZ2
 
 | | CRYSTAL STRUCTURE OF 3-MBT REPEATS OF LETHAL (3) MALIGNANT BRAIN TUMOR (NATIVE-II) AT 1.55 ANGSTROM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
1OZ3
 
 | | Crystal Structure of 3-MBT repeats of lethal (3) malignant Brain Tumor (Native-I) at 1.85 angstrom | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
1OZ6
 
 | | X-ray structure of acidic phospholipase A2 from Indian saw-scaled viper (Echis carinatus) with a potent platelet aggregation inhibitory activity | | Descriptor: | CALCIUM ION, phospholipase A2 | | Authors: | Jasti, J, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 2003-04-08 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an acidic phospholipase A2 from Indian saw-scaled viper (Echis carinatus) at 2.6 A resolution reveals a novel intermolecular interaction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OZ7
 
 | | Crystal structure of Echicetin from the venom of Indian saw-scaled viper (Echis carinatus) at 2.4 resolution | | Descriptor: | echicetin A-chain, echicetin B-chain | | Authors: | Jasti, J, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 2003-04-08 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of echicetin from Echis carinatus (Indian saw-scaled viper) at 2.4A resolution.
J.Mol.Biol., 335, 2004
|
|
1OZ9
 
 | | Crystal structure of AQ_1354, a hypothetical protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_1354 | | Authors: | Oganesyan, V, Busso, D, Brandsen, J, Chen, S, Jancarik, J, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OZA
 
 | |
1OZB
 
 | | Crystal Structure of SecB complexed with SecA C-terminus | | Descriptor: | Preprotein translocase secA subunit, Protein-export protein secB, ZINC ION | | Authors: | Zhou, J, Xu, Z. | | Deposit date: | 2003-04-08 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural determinants of SecB recognition by SecA in bacterial protein translocation
NAT.STRUCT.BIOL., 10, 2003
|
|
1OZF
 
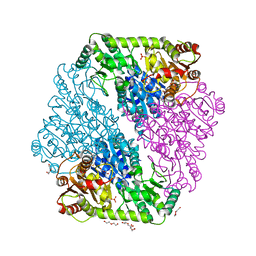 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactors | | Descriptor: | Acetolactate synthase, catabolic, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1OZG
 
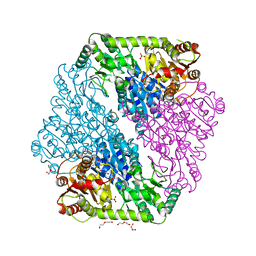 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate | | Descriptor: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1OZH
 
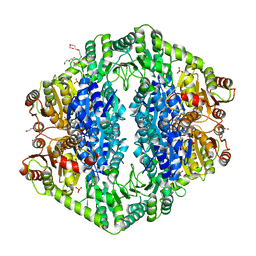 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. | | Descriptor: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1OZM
 
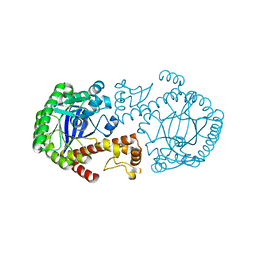 | | Y106F mutant of Z. mobilis TGT | | Descriptor: | Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme
tRNA-guanine transglycosylase and their implications for substrate selectivity,
reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
1OZP
 
 | | Crystal Structure of Rv0819 from Mycobacterium tuberculosis MshD-Mycothiol Synthase Acetyl-Coenzyme A Complex. | | Descriptor: | ACETYL COENZYME *A, hypothetical protein Rv0819 | | Authors: | Vetting, M.W, Roderick, S.L, Yu, M, Blanchard, J.S. | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases.
Protein Sci., 12, 2003
|
|
1OZQ
 
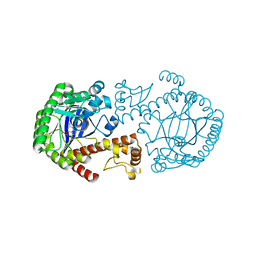 | | CRYSTAL STRUCTURE OF THE MUTATED TRNA-GUANINE TRANSGLYCOSYLASE (TGT)Y106F COMPLEXED WITH PREQ1 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase
and their implications for substrate selectivity, reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
1OZT
 
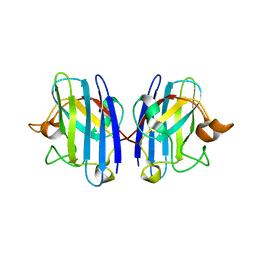 | | Crystal Structure of apo-H46R Familial ALS Mutant human Cu,Zn Superoxide Dismutase (CuZnSOD) to 2.5A resolution | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
1OZU
 
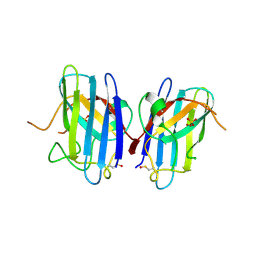 | | Crystal Structure of Familial ALS Mutant S134N of human Cu,Zn Superoxide Dismutase (CuZnSOD) to 1.3A resolution | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
1OZV
 
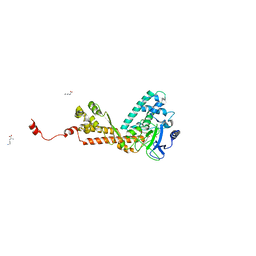 | | Crystal structure of the SET domain of LSMT bound to Lysine and AdoHcy | | Descriptor: | LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | Authors: | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2003-04-09 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
1OZY
 
 | |
1P0B
 
 | | Crystal Structure Of tRNA-Guanine Transglycosylase (TGT) From Zymomonas mobilis Complexed With Archaeosine Precursor, Preq0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase
and their implications for substrate selectivity, reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
1P0C
 
 | | Crystal Structure of the NADP(H)-Dependent Vertebrate Alcohol Dehydrogenase (ADH8) | | Descriptor: | GLYCEROL, NADP-dependent ALCOHOL DEHYDROGENASE, PHOSPHATE ION, ... | | Authors: | Rosell, A, Valencia, E, Pares, X, Fita, I, Farres, J, Ochoa, W.F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Vertebrate NADP(H)-dependent Alcohol Dehydrogenase (ADH8)
J.Mol.Biol., 330, 2003
|
|
1P0D
 
 | | CRYSTAL STRUCTURE OF ZYMOMONAS MOBILIS tRNA-GUANINE TRANSGLYCOSYLASE (TGT) CRYSTALLISED AT PH 5.5 | | Descriptor: | Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme
tRNA-guanine transglycosylase and their implications for substrate selectivity,
reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
