4XLX
 
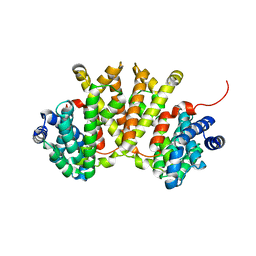 | | Crystal structure of BjKS from Bradyrhizobium japonicum | | Descriptor: | Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4XLY
 
 | | The complex structure of KS-D75C with substrate CPP | | Descriptor: | (2E)-3-methyl-5-[(1R,4aR,8aR)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4XLZ
 
 | | N,N'-diacetylchitobiose deacetylase (SeMet derivative) from Pyrococcus furiosus in the presence of cadmium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM0
 
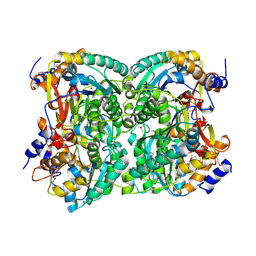 | | N,N'-diacetylchitobiose deacetylase (SeMet derivative) from Pyrococcus furiosus in the absence of cadmium | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM1
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus in the presence of cadmium | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM2
 
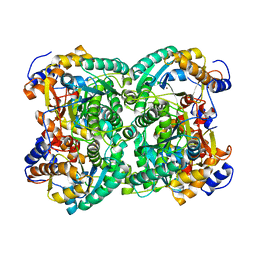 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus in the absence of cadmium | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM3
 
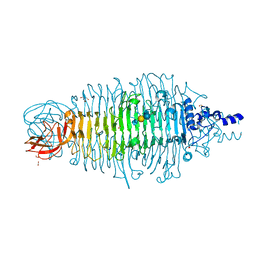 | | Tailspike protein mutant E372A of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XM4
 
 | |
4XM5
 
 | | C. glabrata Slx1. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, ZINC ION | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
4XM6
 
 | | Anthrax toxin lethal factor with ligand-induced binding pocket | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-N~2~-(2-methylpropyl)-D-valinamide, ... | | Authors: | Maize, K.M, De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Ligand-induced expansion of the S1' site in the anthrax toxin lethal factor.
Febs Lett., 589, 2015
|
|
4XM7
 
 | | Anthrax toxin lethal factor with ligand-induced binding pocket | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methoxyphenyl)sulfonyl]-N-hydroxy-N~2~-(2-methylpropyl)-D-valinamide, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2015-01-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand-induced expansion of the S1' site in the anthrax toxin lethal factor.
Febs Lett., 589, 2015
|
|
4XM8
 
 | | Anthrax toxin lethal factor with ligand-induced binding pocket | | Descriptor: | Lethal factor, N-hydroxy-N~2~-{[3-(methoxymethyl)phenyl]sulfonyl}-N~2~-(2-methylpropyl)-D-valinamide, ZINC ION | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2015-01-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand-induced expansion of the S1' site in the anthrax toxin lethal factor.
Febs Lett., 589, 2015
|
|
4XMA
 
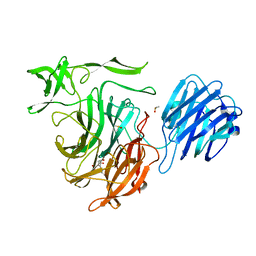 | |
4XMB
 
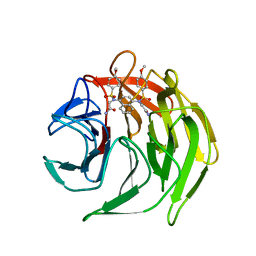 | | Crystal structure of 2,2'-(naphthalene-1,4-diylbis(((4-methoxyphenyl)sulfonyl)azanediyl))diacetamide bound to human Keap1 Kelch domain | | Descriptor: | 2,2'-(naphthalene-1,4-diylbis(((4-methoxyphenyl)sulfonyl)azanediyl))diacetamide, Kelch-like ECH-associated protein 1 | | Authors: | Luciano, J.P, Ryuzoji, A.F, Mesecar, A.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Probing the structural requirements of non-electrophilic naphthalene-based Nrf2 activators.
Eur.J.Med.Chem., 103, 2015
|
|
4XMC
 
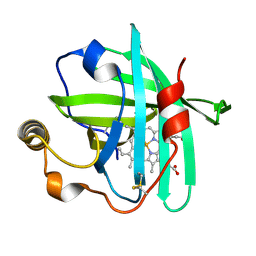 | |
4XMD
 
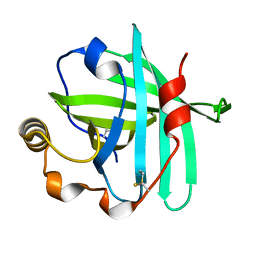 | |
4XME
 
 | |
4XMF
 
 | |
4XMG
 
 | |
4XMH
 
 | |
4XMI
 
 | |
4XMK
 
 | |
4XML
 
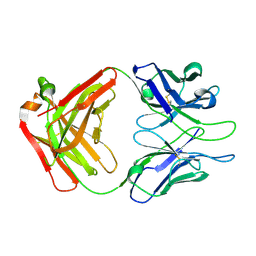 | | Crystal structure of Fab of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Descriptor: | Heavy chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424, Light chain of HIV-1 gp120 V3-specific human monoclonal antibody 2424 | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Functional and Structural Characterization of Human V3-Specific Monoclonal Antibody 2424 with Neutralizing Activity against HIV-1 JRFL.
J.Virol., 89, 2015
|
|
4XMM
 
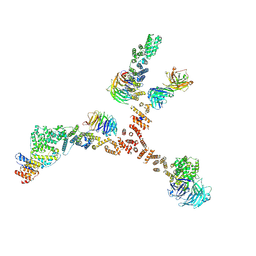 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XMN
 
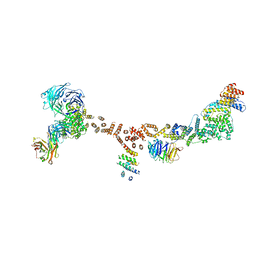 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
