1GK0
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOSPORIN ACYLASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1GK1
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | CEPHALOSPORIN ACYLASE, GLYCEROL | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1GK2
 
 | |
1GK3
 
 | |
1GK4
 
 | | HUMAN VIMENTIN COIL 2B FRAGMENT (CYS2) | | Descriptor: | ACETATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
1GK6
 
 | | Human vimentin coil 2B fragment linked to GCN4 leucine zipper (Z2B) | | Descriptor: | VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
1GK7
 
 | | HUMAN VIMENTIN COIL 1A FRAGMENT (1A) | | Descriptor: | SULFATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
1GK8
 
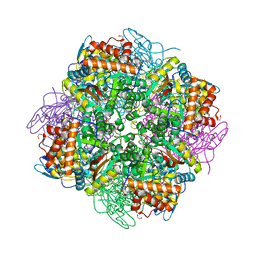 | | Rubisco from Chlamydomonas reinhardtii | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Taylor, T.C. | | Deposit date: | 2001-08-09 | | Release date: | 2001-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | First Crystal Structure of Rubisco from a Green Alga, Chlamydomonas Reinhardtii.
J.Biol.Chem., 276, 2001
|
|
1GK9
 
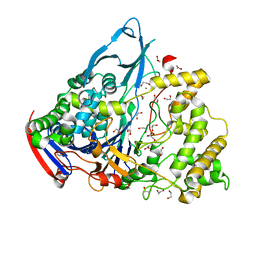 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GKA
 
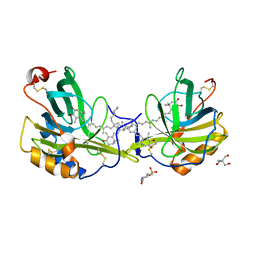 | | The molecular basis of the coloration mechanism in lobster shell. beta-crustacyanin at 3.2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASTAXANTHIN, ... | | Authors: | Cianci, M, Rizkallah, P.J, Olczak, A, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2001-08-10 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | The Molecular Basis of the Coloration Mechanism in Lobster Shell: Beta -Crustacyanin at 3.2-A Resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GKB
 
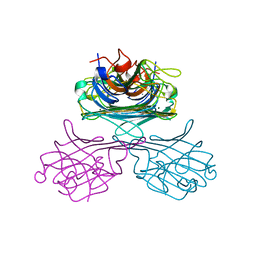 | | CONCANAVALIN A, NEW CRYSTAL FORM | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MAGNESIUM ION, ... | | Authors: | Kantardjieff, K, Rupp, B, Hoechtl, P, Segelke, B. | | Deposit date: | 2001-08-10 | | Release date: | 2001-08-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Concanavalin a in a Dimeric Crystal Form: Revisiting Structural Accuracy and Molecular Flexibility
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GKC
 
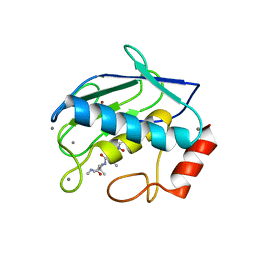 | | MMP9-inhibitor complex | | Descriptor: | 92 KDA TYPE IV COLLAGENASE, CALCIUM ION, N~2~-[(2R)-2-{[formyl(hydroxy)amino]methyl}-4-methylpentanoyl]-N,3-dimethyl-L-valinamide, ... | | Authors: | Rowsell, S, Pauptit, R.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Mmp9 in Complex with a Reverse Hydroxamate Inhibitor
J.Mol.Biol., 319, 2002
|
|
1GKD
 
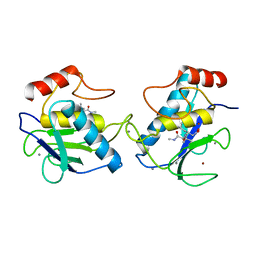 | | MMP9 active site mutant-inhibitor complex | | Descriptor: | 2-AMINO-N,3,3-TRIMETHYLBUTANAMIDE, 2-{[FORMYL(HYDROXY)AMINO]METHYL}-4-METHYLPENTANOIC ACID, 92 KDA TYPE IV COLLAGENASE, ... | | Authors: | Rowsell, S, Pauptit, R.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Mmp9 in Complex with a Reverse Hydroxamate Inhibitor
J.Mol.Biol., 319, 2002
|
|
1GKF
 
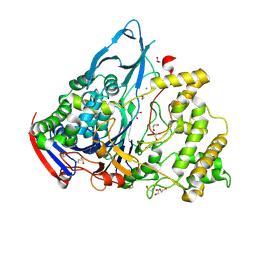 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-13 | | Release date: | 2002-01-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GKH
 
 | | MUTANT K69H OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1GKJ
 
 | |
1GKK
 
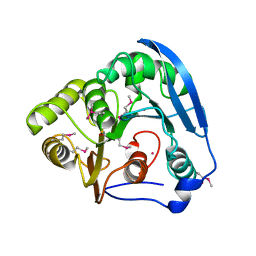 | | Feruloyl esterase domain of XynY from clostridium thermocellum | | Descriptor: | CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, GLYCEROL | | Authors: | Prates, J.A.M, Tarbouriech, N, Charnock, S.J, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-12-13 | | Last modified: | 2011-09-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum Provides Insights Into Substrate Recognition
Structure, 9, 2001
|
|
1GKL
 
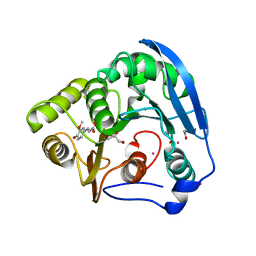 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ACETATE ION, CADMIUM ION, ... | | Authors: | Prates, J.A.M, Tarbouriech, N, Charnock, S.J, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of the feruloyl esterase module of xylanase 10B from Clostridium thermocellum provides insights into substrate recognition.
Structure, 9, 2001
|
|
1GKM
 
 | |
1GKP
 
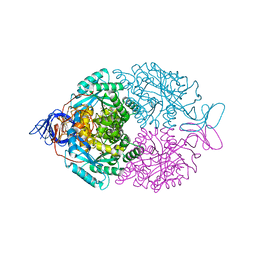 | | D-Hydantoinase (Dihydropyrimidinase) from Thermus sp. in space group C2221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HYDANTOINASE, SULFATE ION, ... | | Authors: | Abendroth, J, Niefind, K, Schomburg, D. | | Deposit date: | 2001-08-20 | | Release date: | 2002-06-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.295 Å) | | Cite: | X-Ray Structure of a Dihydropyrimidinase from Thermus Sp. At 1.3 A Resolution
J.Mol.Biol., 320, 2002
|
|
1GKQ
 
 | |
1GKR
 
 | |
1GKU
 
 | |
1GKX
 
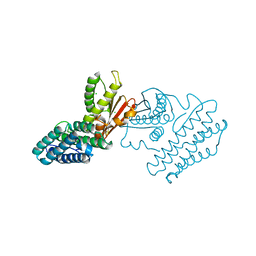 | | Branched-chain alpha-ketoacid dehydrogenase kinase (BCK) | | Descriptor: | CHLORIDE ION, [3-METHYL-2-OXOBUTANOATE DEHYDROGENASE [LIPOAMIDE]] KINASE | | Authors: | Machius, M, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2001-08-21 | | Release date: | 2001-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Rat Bckd Kinase: Nucleotide-Induced Domain Communication in a Mitochondrial Protein Kinase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GKY
 
 | |
