1F5J
 
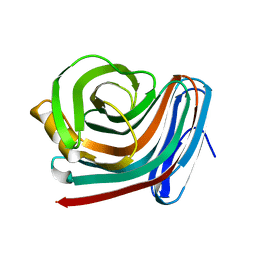 | | CRYSTAL STRUCTURE OF XYNB, A HIGHLY THERMOSTABLE BETA-1,4-XYLANASE FROM DICTYOGLOMUS THERMOPHILUM RT46B.1, AT 1.8 A RESOLUTION | | Descriptor: | BETA-1,4-XYLANASE, SULFATE ION | | Authors: | McCarthy, A.A, Baker, E.N. | | Deposit date: | 2000-07-26 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of XynB, a highly thermostable beta-1,4-xylanase from Dictyoglomus thermophilum Rt46B.1, at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1F5M
 
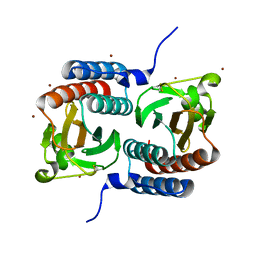 | | STRUCTURE OF THE GAF DOMAIN | | Descriptor: | BROMIDE ION, GAF | | Authors: | Ho, Y.S, Burden, L.M, Hurley, J.H. | | Deposit date: | 2000-06-15 | | Release date: | 2000-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor.
EMBO J., 19, 2000
|
|
1F5O
 
 | |
1F5P
 
 | |
1F5S
 
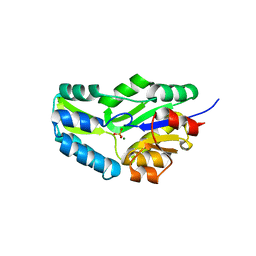 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOSERINE PHOSPHATASE (PSP) | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-06-15 | | Release date: | 2001-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution.
Structure, 9, 2001
|
|
1F5T
 
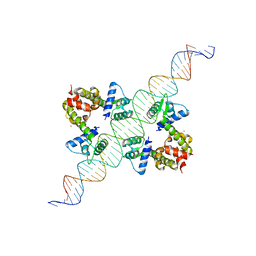 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT) COMPLEXED WITH NICKEL AND DTXR CONSENSUS BINDING SEQUENCE | | Descriptor: | 43MER DNA CONTAINING DXTR CONSENSUS BINDING SEQUENCE, DIPHTHERIA TOXIN REPRESSOR, NICKEL (II) ION | | Authors: | Chen, S, White, A, Love, J, Murphy, J.R, Ringe, D. | | Deposit date: | 2000-06-15 | | Release date: | 2000-09-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Methyl groups of thymine bases are important for nucleic acid recognition by DtxR.
Biochemistry, 39, 2000
|
|
1F5V
 
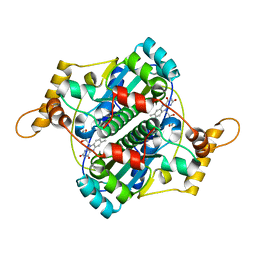 | | STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF A FLAVOPROTEIN FROM ESCHERICHIA COLI THAT REDUCES NITROCOMPOUNDS. ALTERATION OF PYRIDINE NUCLEOTIDE BINDING BY A SINGLE AMINO ACID SUBSTITUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NADPH NITROREDUCTASE | | Authors: | Kobori, T, Sasaki, H, Lee, W.C, Zenno, S, Saigo, K, Murphy, M.E.P, Tanokura, M. | | Deposit date: | 2000-06-17 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.
J.Biol.Chem., 276, 2001
|
|
1F5W
 
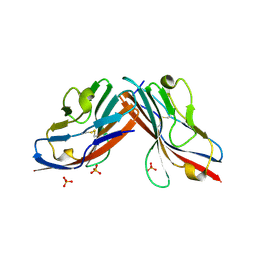 | | DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN | | Descriptor: | COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR, SULFATE ION | | Authors: | van Raaij, M.J, Chouin, E, van der Zandt, H, Bergelson, J.M, Cusack, S. | | Deposit date: | 2000-06-18 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution.
Structure Fold.Des., 8, 2000
|
|
1F60
 
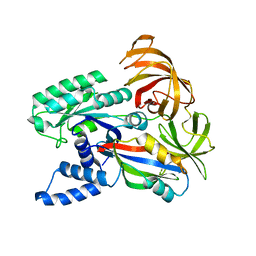 | | CRYSTAL STRUCTURE OF THE YEAST ELONGATION FACTOR COMPLEX EEF1A:EEF1BA | | Descriptor: | ELONGATION FACTOR EEF1A, ELONGATION FACTOR EEF1BA | | Authors: | Andersen, G.R, Pedersen, L, Valente, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2000-06-19 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha.
Mol.Cell, 6, 2000
|
|
1F69
 
 | |
1F6B
 
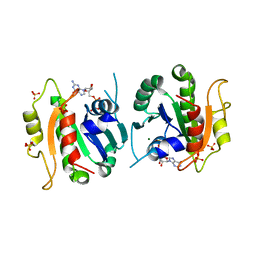 | | CRYSTAL STRUCTURE OF SAR1-GDP COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SAR1, ... | | Authors: | Huang, M, Wilson, I.A, Balch, W.E. | | Deposit date: | 2000-06-21 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export.
J.Cell Biol., 155, 2001
|
|
1F6C
 
 | |
1F6D
 
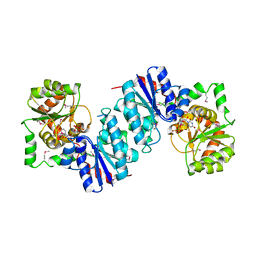 | | THE STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE FROM E. COLI. | | Descriptor: | CHLORIDE ION, SODIUM ION, UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE, ... | | Authors: | Campbell, R.E, Mosimann, S.C, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases.
Biochemistry, 39, 2000
|
|
1F6E
 
 | |
1F6I
 
 | |
1F6J
 
 | |
1F6K
 
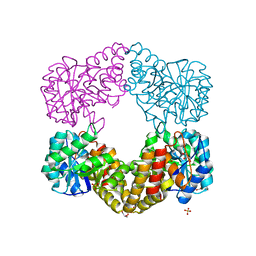 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II | | Descriptor: | GLYCEROL, N-ACETYLNEURAMINATE LYASE, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F6M
 
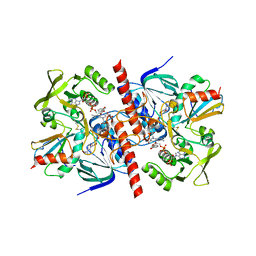 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THIOREDOXIN REDUCTASE, THIOREDOXIN, AND THE NADP+ ANALOG, AADP+ | | Descriptor: | 3-AMINOPYRIDINE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN 1, ... | | Authors: | Lennon, B.W, Williams Jr, C.H, Ludwig, M.L. | | Deposit date: | 2000-06-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase.
Science, 289, 2000
|
|
1F6P
 
 | |
1F6W
 
 | |
1F6Y
 
 | |
1F73
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM III IN COMPLEX WITH SIALIC ACID ALDITOL | | Descriptor: | 2,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDO NONANOIC ACID, GLYCEROL, N-ACETYL NEURAMINATE LYASE | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-25 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F74
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II COMPLEXED WITH 4-DEOXY-SIALIC ACID | | Descriptor: | 6,7,8,9-TETRAHYDROXY-5-METHYLCARBOXAMIDO-2-OXONONANOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-26 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F76
 
 | | ESCHERICHIA COLI DIHYDROOROTATE DEHYDROGENASE | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, FORMIC ACID, ... | | Authors: | Norager, S, Jensen, K.F, Bjornberg, O, Larsen, S. | | Deposit date: | 2000-06-26 | | Release date: | 2002-10-16 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E. coli Dihydroorotate Dehydrogenase Reveals Structural and Functional Distinction between different classes of
dihydroorotate dehydrogenases
Structure, 10, 2002
|
|
1F7B
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II IN COMPLEX WITH 4-OXO-SIALIC ACID | | Descriptor: | 4,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDONONANOIC ACID, 6,7,8,9-TETRAHYDROXY-5-METHYLCARBOXAMIDO-4-OXONONANOIC ACID, CHLORIDE ION, ... | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-26 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
