1S68
 
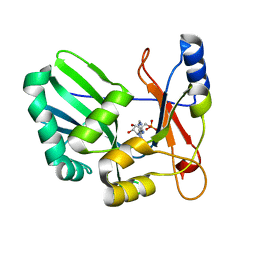 | | Structure and Mechanism of RNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase 2 | | Authors: | Ho, C.K, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of RNA ligase.
Structure, 12, 2004
|
|
1S69
 
 | | The X-ray structure of the cyanobacteria Synechocystis hemoglobin "cyanoglobin" with cyanide ligand | | Descriptor: | CITRATE ANION, CYANIDE ION, Cyanoglobin, ... | | Authors: | Trent III, J.T, Kundu, S, Hoy, J.A, Hargrove, M.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic analysis of synechocystis cyanoglobin reveals the structural changes accompanying ligand binding in a hexacoordinate hemoglobin.
J.Mol.Biol., 341, 2004
|
|
1S6A
 
 | | The X-ray structure of the cyanobacteria Synechocystis hemoglobin "cyanoglobin" with azide ligand | | Descriptor: | AZIDE ION, CITRATE ANION, Cyanoglobin, ... | | Authors: | Trent III, J.T, Kundu, S, Hoy, J.A, Hargrove, M.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic analysis of synechocystis cyanoglobin reveals the structural changes accompanying ligand binding in a hexacoordinate hemoglobin.
J.Mol.Biol., 341, 2004
|
|
1S6C
 
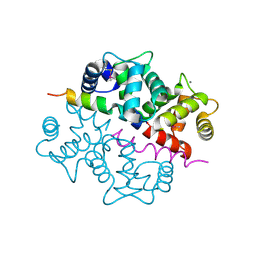 | | Crystal structure of the complex between KChIP1 and Kv4.2 N1-30 | | Descriptor: | CALCIUM ION, Kv4 potassium channel-interacting protein KChIP1b, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhou, W, Qian, Y, Kunjilwar, K, Pfaffinger, P.J, Choe, S. | | Deposit date: | 2004-01-23 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the functional interaction of KChIP1 with Shal-type K(+) channels.
Neuron, 41, 2004
|
|
1S6F
 
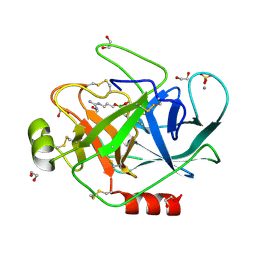 | | PORCINE TRYPSIN COVALENT COMPLEX WITH BORATE AND GUANIDINE-3 INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-23 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S6H
 
 | | PORCINE TRYPSIN COMPLEXED WITH GUANIDINE-3-PROPANOL INHIBITOR | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-23 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S6P
 
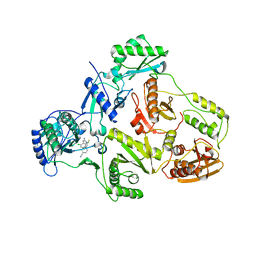 | | CRYSTAL STRUCTURE OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R100943 | | Descriptor: | 1-(4-CYANO-PHENYL)-3-[2-(2,6-DICHLORO-PHENYL)-1-IMINO-ETHYL]-THIOUREA, MAGNESIUM ION, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1S6Q
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S6R
 
 | | 908R class c beta-lactamase bound to iodo-acetamido-phenyl boronic acid | | Descriptor: | 4-IODO-ACETAMIDO PHENYLBORONIC ACID, beta-lactamase | | Authors: | Wouters, J. | | Deposit date: | 2004-01-27 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of Enterobacter cloacae 908R class C beta-lactamase bound to iodo-acetamido-phenyl boronic acid, a transition-state analogue.
Cell.Mol.Life Sci., 60, 2003
|
|
1S6V
 
 | | Structure of a cytochrome c peroxidase-cytochrome c site specific cross-link | | Descriptor: | Cytochrome c peroxidase, mitochondrial, Cytochrome c, ... | | Authors: | Guo, M, Bhaskar, B, Li, H, Barrows, T.P, Poulos, T.L. | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and characterization of a cytochrome c peroxidase-cytochrome c site-specific cross-link
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S6Y
 
 | | 2.3A crystal structure of phospho-beta-glucosidase | | Descriptor: | 6-phospho-beta-glucosidase | | Authors: | Tereshko, V, Dementieva, I, Kim, Y, Collat, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3A CRYSTAL STRUCTURE OF PHOSPHO-BETA-GLUCOSIDASE, licH Gene Product from BACILLUS STEAROTHERMOPHILUS
To be Published
|
|
1S6Z
 
 | | Enhanced Green Fluorescent Protein Containing the Y66L Substitution | | Descriptor: | CHLORIDE ION, green fluorescent protein | | Authors: | Rosenow, M.A, Huffman, H.A, Phail, M.E, Wachter, R.M. | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Y66L Variant of Green Fluorescent Protein Supports a Cyclization-Oxidation-Dehydration Mechanism for Chromophore Maturation(,).
Biochemistry, 43, 2004
|
|
1S72
 
 | | REFINED CRYSTAL STRUCTURE OF THE HALOARCULA MARISMORTUI LARGE RIBOSOMAL SUBUNIT AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10e, 50S ribosomal protein L11P, ... | | Authors: | Klein, D.J, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Roles of Ribosomal Proteins in the Structure, Assembly and Evolution of the Large Ribosomal Subunit
J.Mol.Biol., 340, 2004
|
|
1S73
 
 | | Crystal Structure of Mesopone Cytochrome c Peroxidase (R-isomer) [MpCcP-R] | | Descriptor: | Cytochrome c peroxidase, mitochondrial, FE-(4-MESOPORPHYRINONE)-R-ISOMER | | Authors: | Bhaskar, B, Immoos, C.E, Sulc, F, Choen, M.S, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2004-01-28 | | Release date: | 2005-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structures of reduced, resting and NO-bound states of mesopone cytochorme c peroxidase (MpCcP) (R-isomer)
To be Published
|
|
1S76
 
 | | T7 RNA polymerase alpha beta methylene ATP elongation complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (5'-D(P*GP*CP*CP*GP*TP*GP*CP*GP*CP*AP*TP*TP*CP*GP*CP*CP*GP*TP*GP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*GP*TP*TP*GP*CP*GP*CP*AP*CP*GP*GP*C)-3'), ... | | Authors: | Yin, Y.W, Steitz, T.A. | | Deposit date: | 2004-01-29 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | The structural mechanism of translocation and helicase activity in T7 RNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1S77
 
 | | T7 RNAP product pyrophosphate elongation complex | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*TP*GP*CP*GP*CP*AP*TP*TP*CP*GP*CP*CP*GP*TP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*TP*AP*CP*GP*TP*TP*GP*CP*GP*CP*AP*CP*GP*GP*C)-3'), DNA-directed RNA polymerase, ... | | Authors: | Yin, Y.W, Steitz, T.A. | | Deposit date: | 2004-01-29 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The structural mechanism of translocation and helicase activity in T7 RNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1S78
 
 | | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, ... | | Authors: | Franklin, M.C, Carey, K.D, Vajdos, F.F, Leahy, D.J, de Vos, A.M, Sliwkowski, M.X. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex.
Cancer Cell, 5, 2004
|
|
1S7C
 
 | | Crystal structure of MES buffer bound form of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase A, SULFATE ION | | Authors: | Shin, D.H, Thor, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-29 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of MES buffer bound form of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli
To be Published
|
|
1S7D
 
 | |
1S7F
 
 | | RimL- Ribosomal L7/L12 alpha-N-protein acetyltransferase crystal form I (apo) | | Descriptor: | CHLORIDE ION, MALONIC ACID, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
1S7G
 
 | | Structural Basis for the Mechanism and Regulation of Sir2 Enzymes | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, HEXAETHYLENE GLYCOL, ... | | Authors: | Avalos, J.L, Boeke, J.D, Wolberger, C. | | Deposit date: | 2004-01-29 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and regulation of sir2 enzymes
Mol.Cell, 13, 2004
|
|
1S7H
 
 | | Structural Genomics, 2.2A crystal structure of protein YKOF from Bacillus subtilis | | Descriptor: | ykoF | | Authors: | Zhang, R, Lezondra, L, Moy, S, Dementieva, I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-29 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2A crystal structure of protein YKOF from Bacillus subtilis
To be Published
|
|
1S7I
 
 | | 1.8 A Crystal Structure of a Protein of Unknown Function PA1349 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA1349 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-29 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8A crystal structure of a hypothetical protein PA1349 from Pseudomonas aeruginosa
To be Published
|
|
1S7J
 
 | |
1S7K
 
 | |
