1OC9
 
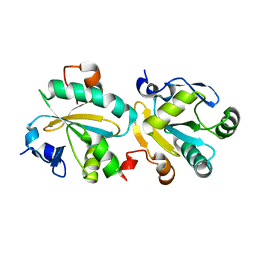 | | TRYPAREDOXIN II FROM C.FASCICULATA solved by MR | | Descriptor: | TRYPAREDOXIN II | | Authors: | Leonard, G.A, Micossi, E, Hunter, W.N. | | Deposit date: | 2003-02-07 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
1OCB
 
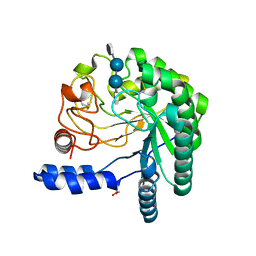 | | Structure of the wild-type cellobiohydrolase Cel6A from Humicolas insolens in complex with a fluorescent substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-amino-4-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-methyl 4-thio-beta-D-glucopyranoside, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1OCE
 
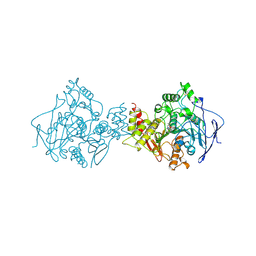 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH MF268 | | Descriptor: | ACETYLCHOLINESTERASE, CIS-2,6-DIMETHYLMORPHOLINOOCTYLCARBAMYLESEROLINE | | Authors: | Bartolucci, C, Perola, E, Cellai, L, Brufani, M, Lamba, D. | | Deposit date: | 1998-06-12 | | Release date: | 1999-05-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | "Back door" opening implied by the crystal structure of a carbamoylated acetylcholinesterase.
Biochemistry, 38, 1999
|
|
1OCJ
 
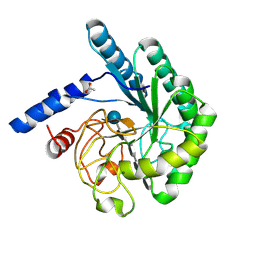 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a THIOPENTASACCHARIDE at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1OCN
 
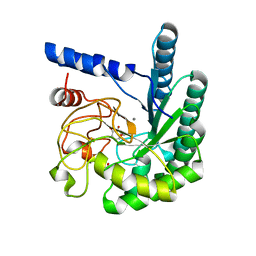 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a cellobio-derived isofagomine at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Distortion of a Cellobio-Derived Isofagomine Highlights the Potential Conformational Itinerary of Inverting Beta-Glucosidases
Chem.Commun.(Camb.), 21, 2003
|
|
1OCQ
 
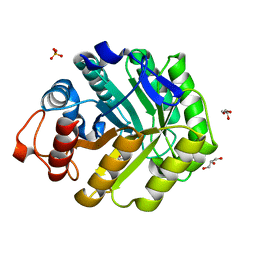 | | COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDOGLUCANASE 5A, GLYCEROL, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Direct Observation of the Protonation State of an Imino Sugar Glycosidase Inhibitor Upon Binding
J.Am.Chem.Soc., 125, 2003
|
|
1OCS
 
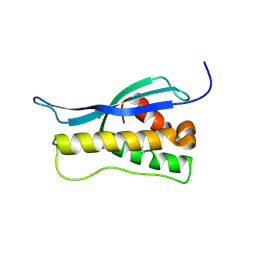 | | Crystal structure of the yeast PX-doamin protein Grd19p (sorting nexin3) complexed to phosphatidylinosytol-3-phosphate. | | Descriptor: | GLYCEROL, SORTING NEXIN GRD19 | | Authors: | Zhou, C.Z, Li De La Sierra-Gallay, I, Cheruel, S, Collinet, B, Minard, P, Blondeau, K, Henkes, G, Aufrere, R, Leulliot, N, Graille, M, Sorel, I, Savarin, P, De La Torre, F, Poupon, A, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2003-02-10 | | Release date: | 2003-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the Yeast Phox Homology (Px) Protein Grd19P (Sorting Nexin 3) Complexed to Phosphatidylinositol-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1OCU
 
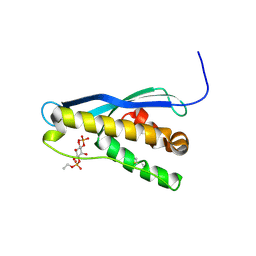 | | Crystal structure of the yeast PX-domain protein Grd19p (sorting nexin 3) complexed to phosphatidylinosytol-3-phosphate. | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, SORTING NEXIN | | Authors: | Zhou, C.Z, Li de La Sierra-Gallay, I, Cheruel, S, Collinet, B, Minard, P, Blondeau, K, Henkes, G, Aufrere, R, Leulliot, N, Graille, M, Sorel, I, Savarin, P, de la Torre, F, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-02-10 | | Release date: | 2003-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate.
J. Biol. Chem., 278, 2003
|
|
1OCV
 
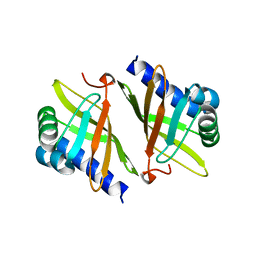 | |
1OCW
 
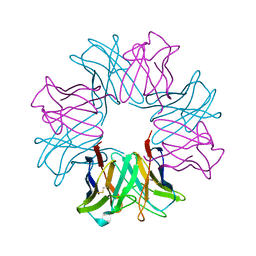 | |
1OCX
 
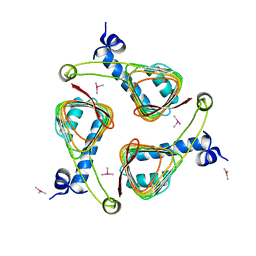 | | E. coli maltose-O-acetyltransferase | | Descriptor: | MALTOSE O-ACETYLTRANSFERASE, TRIMETHYL LEAD ION | | Authors: | Lo Leggio, L, Dal Degan, F, Poulsen, P, Larsen, S. | | Deposit date: | 2003-02-11 | | Release date: | 2003-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure and Specificity of Escherichia Coli Maltose Acetyltransferase Give New Insight Into the Laca Family of Acyltransferases.
Biochemistry, 42, 2003
|
|
1OCY
 
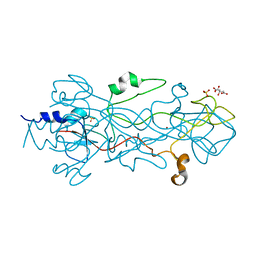 | | Structure of the receptor-binding domain of the bacteriophage T4 short tail fibre | | Descriptor: | BACTERIOPHAGE T4 SHORT TAIL FIBRE, CITRIC ACID, SULFATE ION, ... | | Authors: | Thomassen, E, Gielen, G, Schuetz, M, Miller, S, van Raaij, M.J. | | Deposit date: | 2003-02-11 | | Release date: | 2003-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Receptor-Binding Domain of the Bacteriophage T4 Short Tail Fibre Reveals a Knitted Trimeric Metal-Binding Fold
J.Mol.Biol., 331, 2003
|
|
1OD0
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
1OD1
 
 | | Endothiapepsin PD135,040 complex | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-12 | | Last modified: | 2012-11-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The Structure of Endothiapepsin Complexed with the Gem-Diol Inhibitor Pd-135,040 at 1.37 A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OD2
 
 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL COENZYME *A, ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
1OD3
 
 | | Structure of CSCBM6-3 From Clostridium stercorarium in complex with laminaribiose | | Descriptor: | ACETIC ACID, CALCIUM ION, PUTATIVE XYLANASE, ... | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
1OD4
 
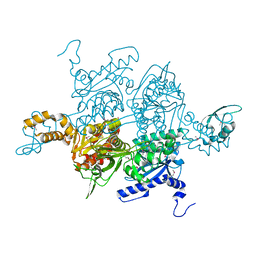 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
1OD5
 
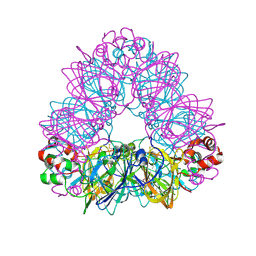 | | Crystal structure of glycinin A3B4 subunit homohexamer | | Descriptor: | CARBONATE ION, GLYCININ, MAGNESIUM ION | | Authors: | Adachi, M, Kanamori, J, Masuda, T, Yagasaki, K, Kitamura, K, Mikami, B, Utsumi, S. | | Deposit date: | 2003-02-13 | | Release date: | 2003-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Soybean 11S Globulin: Glycinin A3B4 Homohexamer
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OD6
 
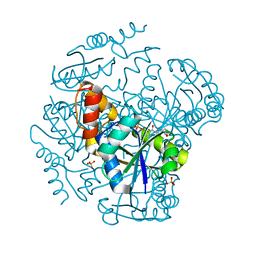 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OD7
 
 | | N-terminal of Sialoadhesin in complex with Me-a-9-N-(naphthyl-2-carbonyl)-amino-9-deoxy-Neu5Ac (NAP compound) | | Descriptor: | ME-A-9-N-(NAPHTHYL-2-CARBONYL)-AMINO-9-DEOXY-NEU5AC, SIALOADHESIN | | Authors: | Zaccai, N.R, Maenaka, K, Maenaka, T, Crocker, P.R, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2003-02-14 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Design of Sialic Acid-Based Siglec Inhibitors and Crystallographic Analysis in Complex with Sialoadhesin
Structure, 11, 2003
|
|
1OD8
 
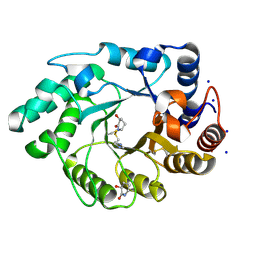 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine lactam | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, SODIUM ION, ... | | Authors: | Gloster, T.M, Roberts, S, Davies, G.J. | | Deposit date: | 2003-02-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A Xylobiose-Derived Isofagomine Lactam Glycosidase Inhibitor Binds as its Amide Tautomer
Chem.Commun.(Camb.), 8, 2003
|
|
1ODB
 
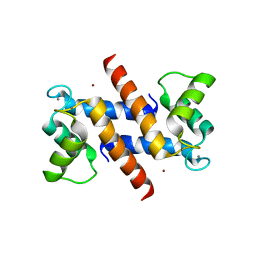 | | THE CRYSTAL STRUCTURE OF HUMAN S100A12 - COPPER COMPLEX | | Descriptor: | CALCIUM ION, CALGRANULIN C, COPPER (II) ION | | Authors: | Moroz, O.V, Antson, A.A, Grist, S.J, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2003-02-15 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the Human S100A12-Copper Complex: Implications for Host-Parasite Defence
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1ODC
 
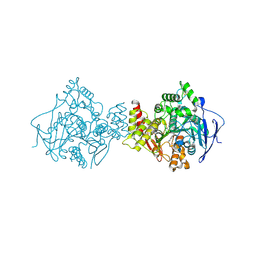 | | STRUCTURE OF ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH N-4'-QUINOLYL-N'-9"-(1",2",3",4"-TETRAHYDROACRIDINYL)-1,8- DIAMINOOCTANE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, N-QUINOLIN-4-YL-N'-(1,2,3,4-TETRAHYDROACRIDIN-9-YL)OCTANE-1,8-DIAMINE | | Authors: | Wong, D.M, Greenblatt, H.M, Carlier, P.R, Han, Y.-F, Pang, Y.-P, Silman, I, Sussman, J.L. | | Deposit date: | 2003-02-15 | | Release date: | 2005-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complexes of Alkylene-Linked Tacrine Dimers with Torpedo Californica Acetylcholinesterase: Binding of Bis(5)-Tacrine Produces a Dramatic Rearrangement in the Active-Site Gorge.
J.Med.Chem., 49, 2006
|
|
1ODE
 
 | |
1ODF
 
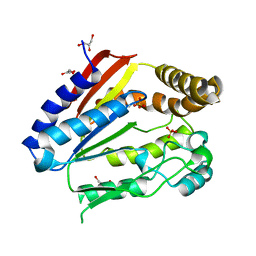 | | Structure of YGR205w protein. | | Descriptor: | GLYCEROL, HYPOTHETICAL 33.3 KDA PROTEIN IN ADE3-SER2 INTERGENIC REGION, SULFATE ION | | Authors: | Li De La Sierra-Gallay, I, Van Tilbeurgh, H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ygr205W Protein from Saccharomyces Cerevisiae: Close Structural Resemblance to E.Coli Pantothenate Kinase
Proteins: Struct.,Funct., Genet., 54, 2004
|
|
