1NRZ
 
 | |
1NS0
 
 | |
1NS2
 
 | |
1NS4
 
 | |
1NS5
 
 | | X-RAY STRUCTURE OF YBEA FROM E.COLI. NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER45 | | Descriptor: | Hypothetical protein ybeA | | Authors: | Benach, J, Shen, J, Rost, B, Xiao, R, Acton, T, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of YBEA from E. coli
To be Published
|
|
1NS6
 
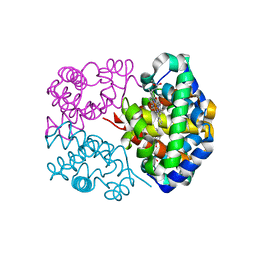 | |
1NS7
 
 | |
1NS8
 
 | |
1NS9
 
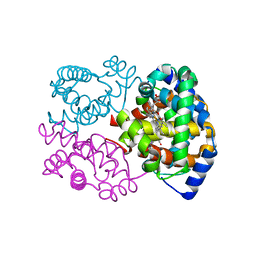 | |
1NSG
 
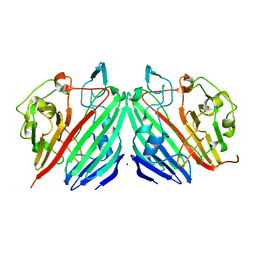
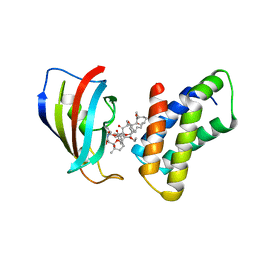 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FKBP-RAPAMYCIN ASSOCIATED PROTEIN (FRAP) | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1NSK
 
 | |
1NSL
 
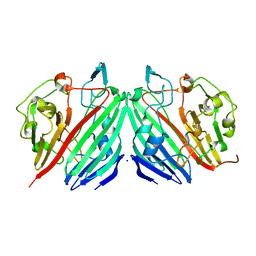
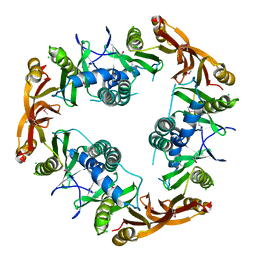 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
1NSM
 
 | |
1NSN
 
 | |
1NSR
 
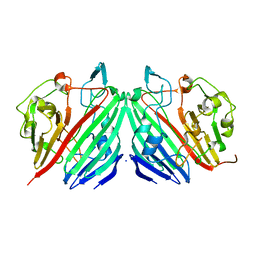 | |
1NSS
 
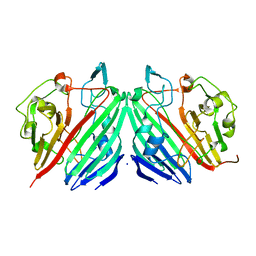 | |
1NSU
 
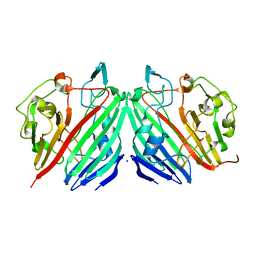 | |
1NSV
 
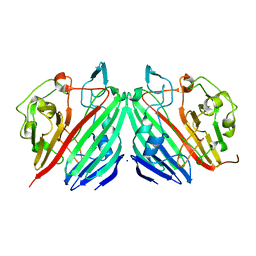 | |
1NSW
 
 | | The Crystal Structure of the K18G Mutant of the thioredoxin from Alicyclobacillus acidocaldarius | | Descriptor: | THIOREDOXIN | | Authors: | Bartolucci, S, De Simone, G, Galdiero, S, Improta, R, Menchise, V, Pedone, C, Pedone, E, Saviano, M. | | Deposit date: | 2003-01-28 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
J.Bacteriol., 185, 2003
|
|
1NSX
 
 | |
1NSZ
 
 | |
1NT0
 
 | | Crystal structure of the CUB1-EGF-CUB2 region of MASP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Feinberg, H, Uitdehaag, J.C.M, Davies, J.M, Wallis, R, Drickamer, K, Weis, W.I. | | Deposit date: | 2003-01-28 | | Release date: | 2003-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CUB1-EGF-CUB2 region of mannose-binding protein associated serine protease-2
Embo J., 22, 2003
|
|
1NT2
 
 | | CRYSTAL STRUCTURE OF FIBRILLARIN/NOP5P COMPLEX | | Descriptor: | Fibrillarin-like pre-rRNA processing protein, S-ADENOSYLMETHIONINE, conserved hypothetical protein | | Authors: | Aittaleb, M, Rashid, R, Chen, Q, Palmer, J.R, Daniels, C.J, Li, H. | | Deposit date: | 2003-01-28 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of archaeal box C/D sRNP core proteins.
Nat.Struct.Biol., 10, 2003
|
|
1NT8
 
 | | Structural Characterisation of the Holliday junction formed by the sequence CCGGTACCGG at 2.00 A | | Descriptor: | 5'-d(CpCpGpGpTpApCpCpGpG)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Texieira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-01-29 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of two Holliday Junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1NTA
 
 | | 2.9 A crystal structure of Streptomycin RNA-aptamer | | Descriptor: | 5'-R(*CP*GP*GP*CP*AP*CP*CP*AP*CP*GP*GP*UP*CP*GP*GP*AP*UP*C)-3', 5'-R(*GP*GP*AP*UP*CP*GP*CP*AP*UP*UP*UP*GP*GP*AP*CP*UP*UP*CP*UP*GP*CP*C)-3', BARIUM ION, ... | | Authors: | Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2003-01-29 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Encapsulating Streptomycin within a small 40-mer RNA
CHEM.BIOL., 10, 2003
|
|
