1N7H
 
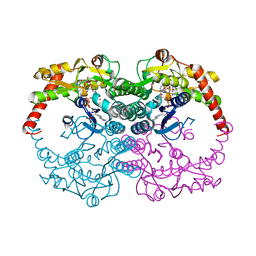 | | Crystal Structure of GDP-mannose 4,6-dehydratase ternary complex with NADPH and GDP | | Descriptor: | GDP-D-mannose-4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mulichak, A.M, Bonin, C.P, Reiter, W.-D, Garavito, R.M. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the MUR1 GDP-mannose
4,6-dehydratase from A. thaliana:
Implications for ligand binding and specificity.
Biochemistry, 41, 2002
|
|
1N7I
 
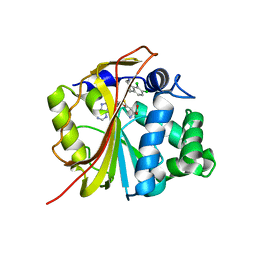 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and the inhibitor LY134046 | | Descriptor: | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO-1H-BENZO[C]AZEPINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1N7J
 
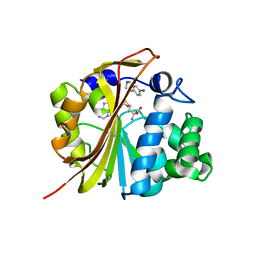 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and an iodinated inhibitor | | Descriptor: | 7-IODO-1,2,3,4-TETRAHYDRO-ISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1N7M
 
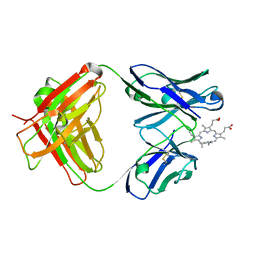 | | Germline 7G12 with N-methylmesoporphyrin | | Descriptor: | Germline Metal Chelatase Catalytic Antibody, chain H, chain L, ... | | Authors: | Yin, J, Andryski, S.E, Beuscher IV, A.E, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2002-11-15 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for substrate strain in antibody catalysis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N7N
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W292A Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7O
 
 | | Streptococcus pneumoniae Hyaluronate Lyase F343V Mutant | | Descriptor: | hyaluronidase | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7P
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W292A/F343V Double Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7Q
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W291A/W292A Double Mutant complex with hyaluronan hexasacchride | | Descriptor: | HYALURONIDASE, beta-D-galactopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7R
 
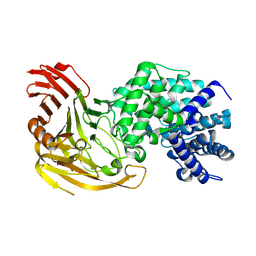 | | Streptococcus pneumoniae Hyaluronate Lyase W291A/W292A/F343V Mutant complex with hexasaccharide hyaluronan | | Descriptor: | HYALURONIDASE, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7S
 
 | | High Resolution Structure of a Truncated Neuronal SNARE Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, SNAP-25A, ... | | Authors: | Ernst, J.A, Brunger, A.T. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High Resolution Structure, Stability, and Synaptotagmin Binding of a Truncated Neuronal SNARE Complex
J.Biol.Chem., 278, 2003
|
|
1N7U
 
 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM I | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
1N7V
 
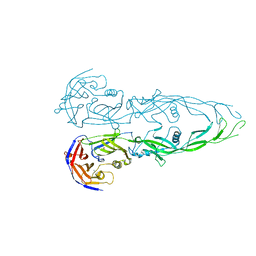 | | THE RECEPTOR-BINDING PROTEIN P2 OF BACTERIOPHAGE PRD1: CRYSTAL FORM III | | Descriptor: | ACETATE ION, Adsorption protein P2, CALCIUM ION | | Authors: | Xu, L, Benson, S.D, Butcher, S.J, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-11-18 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Receptor Binding Protein P2 of PRD1, a
Virus Targeting Antibiotic-Resistant Bacteria,
Has a Novel Fold Suggesting Multiple Functions.
Structure, 11, 2003
|
|
1N7W
 
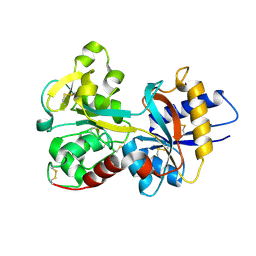 | | Crystal Structure of Human Serum Transferrin, N-Lobe L66W mutant | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, MacGillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-18 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Position of Arginine 124 Controls the Rate of Iron Release from the N-lobe of Human Serum Transferrin. A Structural Study
J.Biol.Chem., 278, 2003
|
|
1N7X
 
 | | HUMAN SERUM TRANSFERRIN, N-LOBE Y45E MUTANT | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, Macgillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-18 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | THE POSITION OF ARGININE 124 CONTROLS THE RATE OF IRON RELEASE FROM THE N-LOBE OF HUMAN SERUM TRANSFERRIN. A STRUCTURAL STUDY
J.Biol.Chem., 278, 2003
|
|
1N7Y
 
 | | STREPTAVIDIN MUTANT N23E AT 1.96A | | Descriptor: | Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-18 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N7Z
 
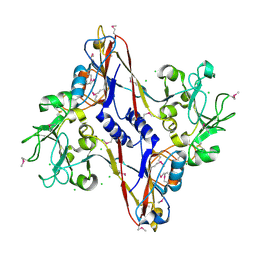 | | Structure and location of gene product 8 in the bacteriophage T4 baseplate | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N80
 
 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N81
 
 | |
1N83
 
 | | Crystal Structure of the complex between the Orphan Nuclear Hormone Receptor ROR(alpha)-LBD and Cholesterol | | Descriptor: | CHOLESTEROL, Nuclear receptor ROR-alpha | | Authors: | Kallen, J.A, Schlaeppi, J.M, Bitsch, F, Geisse, S, Geiser, M, Delhon, I, Fournier, B. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure of hROR(alpha) LBD at 1.63A: Structural and Functional data that Cholesterol or a Cholesterol derivative is the natural ligand of ROR(alpha)
Structure, 10, 2002
|
|
1N84
 
 | | HUMAN SERUM TRANSFERRIN, N-LOBE | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, Macgillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | THE POSITION OF ARGININE 124 CONTROLS THE RATE OF IRON RELEASE FROM THE N-LOBE OF HUMAN SERUM TRANSFERRIN. A STRUCTURAL STUDY
J.Biol.Chem., 278, 2003
|
|
1N8B
 
 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | BROMIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-20 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N8F
 
 | | Crystal structure of E24Q mutant of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Escherichia Coli in complex with Mn2+ and PEP | | Descriptor: | DAHP Synthetase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Shumilin, I.A, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2002-11-20 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The High-Resolution Structure of 3-Deoxy-D-arabino-heptulosonate-7-phosphate
Synthase Reveals a Twist in the Plane of Bound Phosphoenolpyruvate
Biochemistry, 42, 2003
|
|
1N8J
 
 | |
1N8K
 
 | |
1N8N
 
 | | Crystal structure of the Au3+ complex of AphA class B acid phosphatase/phosphotransferase from E. coli at 1.69 A resolution | | Descriptor: | Class B acid phosphatase, GOLD 3+ ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2002-11-21 | | Release date: | 2004-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The first structure of a bacterial class B Acid phosphatase reveals further structural heterogeneity among phosphatases of the haloacid dehalogenase fold.
J.Mol.Biol., 335, 2004
|
|
