1GTF
 
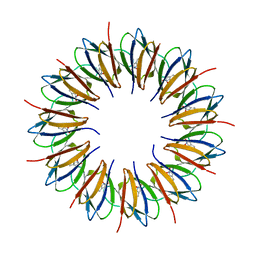 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a 53-nucleotide RNA molecule containing GAGUU repeats | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRP RNA-BINDING ATTENUATION PROTEIN (TRAP), TRYPTOPHAN | | Authors: | Hopcroft, N.H, Wendt, A.L, Gollnick, P, Antson, A.A. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specificity of Trap-RNA Interactions: Crystal Structures of Two Complexes with Different RNA Sequences
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GTH
 
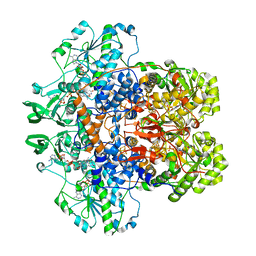 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTK
 
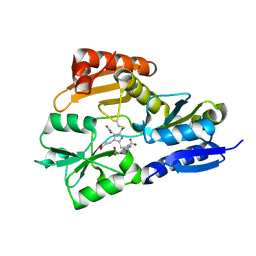 | | Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PORPHOBILINOGEN DEAMINASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 2002-01-16 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Time-Resolved and Static-Ensemble Structural Chemistry of Hydroxymethylbilane Synthase
Faraday Discuss., 122, 2003
|
|
1GTN
 
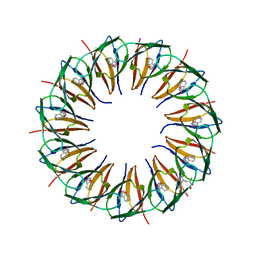 | | Structure of the trp RNA-binding attenuation protein (TRAP) bound to an RNA molecule containing 11 GAGCC repeats | | Descriptor: | (GAGCC)11G 56-NUCLEOTIDE RNA, TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Wendt, A.L, Gollnick, P, Antson, A.A. | | Deposit date: | 2002-01-16 | | Release date: | 2002-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Specificity of Trap-RNA Interactions: Crystal Structures of Two Complexes with Different RNA Sequences
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GTO
 
 | |
1GTT
 
 | | CRYSTAL STRUCTURE OF HPCE | | Descriptor: | 4-HYDROXYPHENYLACETATE DEGRADATION BIFUNCTIONAL ISOMERASE/DECARBOXYLASE, CALCIUM ION | | Authors: | Tame, J.R.H, Namba, K, Dodson, E.J, Roper, D.I. | | Deposit date: | 2002-01-18 | | Release date: | 2002-03-08 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Hpce, a Bifunctional Decarboxylase/Isomerase with a Multifunctional Fold.
Biochemistry, 41, 2002
|
|
1GTU
 
 | |
1GTV
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE-5'-DIPHOSPHATE (TDP) | | Descriptor: | ACETATE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ursby, T, Weik, M, Fioravanti, E, Delarue, M, Goeldner, M, Bourgeois, D. | | Deposit date: | 2002-01-21 | | Release date: | 2002-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cryophotolysis of Caged Compounds: A Technique for Trapping Intermediate States in Protein Crystals
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GTW
 
 | |
1GU2
 
 | | Crystal structure of oxidized cytochrome c'' from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Enguita, F.J, Pohl, E, Rodrigues, A, Santos, H, Carrondo, M.A. | | Deposit date: | 2002-01-22 | | Release date: | 2003-01-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
1GU3
 
 | | CBM4 structure and function | | Descriptor: | ENDOGLUCANASE C, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1GU4
 
 | |
1GU5
 
 | |
1GU7
 
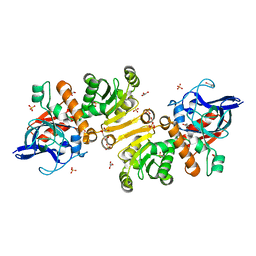 | | Enoyl thioester reductase from Candida tropicalis | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1,MITOCHONDRIAL, GLYCEROL, ... | | Authors: | Airenne, T.T, Torkko, J.M, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1GU8
 
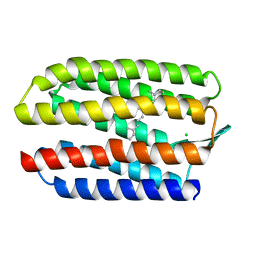 | | SENSORY RHODOPSIN II | | Descriptor: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | Authors: | Edman, K, Royant, A, Nollert, P, Maxwell, C.A, Pebay-Peyroula, E, Navarro, J, Neutze, R, Landau, E.M. | | Deposit date: | 2002-01-24 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Early Structural Rearrangements in the Photocycle of an Integral Membrane Sensory Receptor
Structure, 10, 2002
|
|
1GU9
 
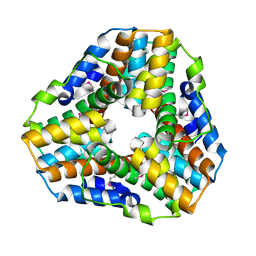 | | Crystal Structure of Mycobacterium tuberculosis Alkylperoxidase AhpD | | Descriptor: | ALKYLHYDROPEROXIDASE D | | Authors: | Nunn, C.M, Djordjevic, S, Hillas, P.J, Nishida, C, Ortiz de Montellano, P.R. | | Deposit date: | 2002-01-24 | | Release date: | 2002-02-14 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Mycobacterium Tuberculosis Alkylhydroperoxidase Ahpd, a Potential Target for Antitubercular Drug Design
J.Biol.Chem., 277, 2002
|
|
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GUF
 
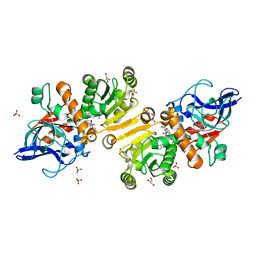 | | Enoyl thioester reductase from Candida tropicalis | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1, MITOCHONDRIAL, ... | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-01-25 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1GUG
 
 | | MopII from Clostridium pasteurianum complexed with tungstate | | Descriptor: | CHLORIDE ION, MOLYBDATE BINDING PROTEIN II, SODIUM ION, ... | | Authors: | Schuettelkopf, A.W, Harrison, J.A, Hunter, W.N. | | Deposit date: | 2002-01-25 | | Release date: | 2002-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Passive Acquisition of Ligand by the Mopii Molbindin from Clostridium Pasteurianum: Structures of Apo and Oxyanion-Bound Forms
J.Biol.Chem., 277, 2002
|
|
1GUH
 
 | | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the MU and PI class enzymes | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1-1, S-BENZYL-GLUTATHIONE | | Authors: | Sinning, I, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 1993-02-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes.
J.Mol.Biol., 232, 1993
|
|
1GUI
 
 | | CBM4 structure and function | | Descriptor: | CALCIUM ION, GLYCEROL, LAMINARINASE 16A, ... | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-27 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1GUL
 
 | | HUMAN GLUTATHIONE TRANSFERASE A4-4 COMPLEX WITH IODOBENZYL GLUTATHIONE | | Descriptor: | GAMMA-GLUTAMYL[S-(2-IODOBENZYL)CYSTEINYL]GLYCINE, Glutathione S-transferase A4 | | Authors: | Bruns, C.M, Hubatsch, I, Ridderstrom, M, Mannervik, B, Tainer, J.A. | | Deposit date: | 1998-06-10 | | Release date: | 1999-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products
J.Mol.Biol., 288, 1999
|
|
1GUM
 
 | | HUMAN GLUTATHIONE TRANSFERASE A4-4 WITHOUT LIGANDS | | Descriptor: | PROTEIN (GLUTATHIONE TRANSFERASE A4-4) | | Authors: | Bruns, C.M, Hubatsch, I, Ridderstrom, M, Mannervik, B, Tainer, J.A. | | Deposit date: | 1998-06-11 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products
J.Mol.Biol., 288, 1999
|
|
1GUN
 
 | |
