1G7N
 
 | |
1G7P
 
 | | CRYSTAL STRUCTURE OF MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND YEAST ALPHA-GLUCOSIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-GLUCOSIDASE P1, ... | | Authors: | Apostolopoulos, V, Yu, M, Corper, A.L, Li, W, McKenzie, I.F, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-11-13 | | Release date: | 2002-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a non-canonical high affinity peptide complexed with MHC class I: a novel use of alternative anchors.
J.Mol.Biol., 318, 2002
|
|
1G7Q
 
 | | CRYSTAL STRUCTURE OF MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND MUC1 VNTR PEPTIDE SAPDTRPA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2 MICROGLOBULIN, ... | | Authors: | Apostolopoulos, V, Yu, M, Corper, A.L, Teyton, L, Pietersz, G.A, McKenzie, I.F, Wilson, I.A. | | Deposit date: | 2000-11-13 | | Release date: | 2002-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a non-canonical low-affinity peptide complexed with MHC class I: a new approach for vaccine design.
J.Mol.Biol., 318, 2002
|
|
1G7Y
 
 | | THE CRYSTAL STRUCTURE OF THE 58KD VEGETATIVE LECTIN FROM THE TROPICAL LEGUME DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, STEM/LEAF LECTIN DB58, ... | | Authors: | Buts, L, Hamelryck, T.W, Loris, R, Dao-Thi, M.-H, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
1G81
 
 | | CHORISMATE LYASE WITH BOUND PRODUCT, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V.L, Howard, A. | | Deposit date: | 2000-11-15 | | Release date: | 2001-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
1G82
 
 | | STRUCTURE OF FIBROBLAST GROWTH FACTOR 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FIBROBLAST GROWTH FACTOR 9, ... | | Authors: | Hecht, H.J, Adar, R, Hofmann, B, Bogin, O, Weich, H, Yayon, A. | | Deposit date: | 2000-11-16 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of fibroblast growth factor 9 shows a symmetric dimer with unique receptor- and heparin-binding interfaces.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G83
 
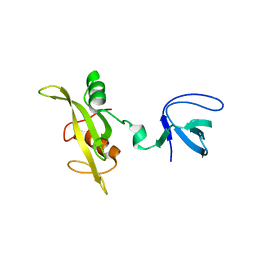 | | CRYSTAL STRUCTURE OF FYN SH3-SH2 | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE FYN | | Authors: | Arold, S.T, Ulmer, T.S, Mulhern, T.D, Werner, J.M, Ladbury, J.E, Campbell, I.D, Noble, M.E.M. | | Deposit date: | 2000-11-16 | | Release date: | 2001-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The role of the Src homology 3-Src homology 2 interface in the regulation of Src kinases.
J.Biol.Chem., 276, 2001
|
|
1G85
 
 | | CRYSTAL STRUCTURE OF BOVINE ODORANT BINDING PROTEIN COMPLEXED WITH IS NATURAL LIGAND | | Descriptor: | (3R)-oct-1-en-3-ol, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-11-16 | | Release date: | 2002-06-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
1G87
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOCELLULASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-16 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1G88
 
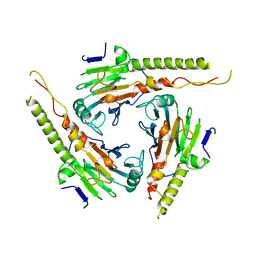 | | S4AFL3ARG515 MUTANT | | Descriptor: | SMAD4 | | Authors: | Chako, B.M, Qin, B, Lam, S.S, Correia, J.J, Lin, K. | | Deposit date: | 2000-11-16 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization.
Nat.Struct.Biol., 8, 2001
|
|
1G8A
 
 | |
1G8F
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1G8G
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE BINARY PRODUCT COMPLEX WITH APS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1G8H
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE TERNARY PRODUCT COMPLEX WITH APS AND PPI | | Descriptor: | ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1G8I
 
 | | CRYSTAL STRUCTURE OF HUMAN FREQUENIN (NEURONAL CALCIUM SENSOR 1) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Bourne, Y, Dannenberg, J, Pollmann, V, Marchot, P, Pongs, O. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1).
J.Biol.Chem., 276, 2001
|
|
1G8J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARSENITE OXIDASE FROM ALCALIGENES FAECALIS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE OXIDASE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ellis, P.J, Conrads, T, Hille, R, Kuhn, P. | | Deposit date: | 2000-11-17 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
Structure, 9, 2001
|
|
1G8K
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARSENITE OXIDASE FROM ALCALIGENES FAECALIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE OXIDASE, ... | | Authors: | Ellis, P.J, Conrads, T, Hille, R, Kuhn, P. | | Deposit date: | 2000-11-17 | | Release date: | 2000-12-13 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
Structure, 9, 2001
|
|
1G8O
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NATIVE BOVINE ALPHA-1,3-GALACTOSYLTRANSFERASE CATALYTIC DOMAIN | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Gastinel, L.N, Bigon, C, Misra, A.K, Hindsgaul, O, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-11-20 | | Release date: | 2001-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
1G8P
 
 | | CRYSTAL STRUCTURE OF BCHI SUBUNIT OF MAGNESIUM CHELATASE | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT | | Authors: | Fodje, M.N, Hansson, A, Hansson, M, Olsen, J.G, Gough, S, Willows, R.D, Al-Karadaghi, S. | | Deposit date: | 2000-11-20 | | Release date: | 2001-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase.
J.Mol.Biol., 311, 2001
|
|
1G8Q
 
 | | CRYSTAL STRUCTURE OF HUMAN CD81 EXTRACELLULAR DOMAIN, A RECEPTOR FOR HEPATITIS C VIRUS | | Descriptor: | CD81 ANTIGEN, EXTRACELLULAR DOMAIN | | Authors: | Kitadokoro, K, Bolognesi, M, Bordo, D, Grandi, G, Galli, G, Petracca, R, Falugi, F. | | Deposit date: | 2000-11-20 | | Release date: | 2001-02-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs.
EMBO J., 20, 2001
|
|
1G8S
 
 | |
1G8T
 
 | | SM ENDONUCLEASE FROM SERATIA MARCENSCENS AT 1.1 A RESOLUTION | | Descriptor: | MAGNESIUM ION, NUCLEASE SM2 ISOFORM, SULFATE ION | | Authors: | Lunin, V.V, Perbandt, M, Betzel, C.H, Mikhailov, A.M. | | Deposit date: | 2000-11-21 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structure of the Serratia marcescens endonuclease at 1.1 A resolution and the enzyme reaction mechanism.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1G8W
 
 | | IMPROVED STRUCTURE OF PHYTOHEMAGGLUTININ-L FROM THE KIDNEY BEAN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LEUCOAGGLUTINATING PHYTOHEMAGGLUTININ, ... | | Authors: | Buts, L, Hamelryck, T.W, Dao-Thi, M, Loris, R, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-21 | | Release date: | 2000-12-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
1G8X
 
 | | STRUCTURE OF A GENETICALLY ENGINEERED MOLECULAR MOTOR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN II HEAVY CHAIN FUSED TO ALPHA-ACTININ 3 | | Authors: | Kliche, W, Fujita-Becker, S, Kollmar, M, Manstein, D.J, Kull, F.J. | | Deposit date: | 2000-11-21 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a genetically engineered molecular motor.
EMBO J., 20, 2001
|
|
1G8Z
 
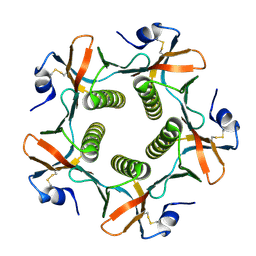 | | HIS57ALA MUTANT OF CHOLERA TOXIN B-PENATMER | | Descriptor: | CHOLERA TOXIN B PROTEIN, beta-D-galactopyranose | | Authors: | Aman, A.T, Fraser, S, Merritt, E.A, Rodigherio, C, Kenny, M. | | Deposit date: | 2000-11-21 | | Release date: | 2001-07-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A mutant cholera toxin B subunit that binds GM1- ganglioside but lacks immunomodulatory or toxic activity.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
