1S28
 
 | | Crystal Structure of AvrPphF ORF1, the Chaperone for the Type III Effector AvrPphF ORF2 from P. syringae | | Descriptor: | ORF1, SULFATE ION | | Authors: | Singer, A.U, Desveaux, D, Betts, L, Chang, J.H, Nimchuk, Z, Grant, S.R, Dangl, J.L, Sondek, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Type III Effector Protein AvrPphF and Its Chaperone Reveal Residues Required for Plant Pathogenesis
Structure, 12, 2004
|
|
1S29
 
 | |
1S2D
 
 | | Purine 2'-Deoxyribosyl complex with arabinoside: Ribosylated Intermediate (AraA) | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-arabinofuranose, ADENINE, Nucleoside 2-deoxyribosyltransferase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S2E
 
 | | BACTERIOPHAGE T4 GENE PRODUCT 9 (GP9), THE TRIGGER OF TAIL CONTRACTION AND THE LONG TAIL FIBERS CONNECTOR, ALTERNATIVE FIT OF THE FIRST 19 RESIDUES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Baseplate structural protein Gp9 | | Authors: | Kostyuchenko, V.A, Navruzbekov, G.A, Kurochkina, L.P, Strelkov, S.V, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2004-01-08 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Bacteriophage T4 Gene Product 9: The Trigger for Tail Contraction
Structure Fold.Des., 7, 1999
|
|
1S2G
 
 | | Purine 2'deoxyribosyltransferase + 2'-deoxyadenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, purine trans deoxyribosylase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S2I
 
 | | Purine 2'deoxyribosyltransferase + bromopurine | | Descriptor: | 6-BROMO-7H-PURINE, purine trans deoxyribosylase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S2J
 
 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
1S2L
 
 | |
1S2M
 
 | |
1S2N
 
 | |
1S2O
 
 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution | | Descriptor: | MAGNESIUM ION, sucrose-phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.L. | | Deposit date: | 2004-01-09 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1S2P
 
 | | The structure and refinement of apocrustacyanin C2 to 1.3A resolution and the search for differences between this protein and the homologous apoproteins A1 and C1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Crustacyanin C2 subunit, SULFATE ION | | Authors: | Habash, J, Helliwell, J.R, Raftery, J, Cianci, M, Rizkallah, P.J, Chayen, N.E, NNeji, G.A, Zakalsky, P.F. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure and refinement of apocrustacyanin C2 to 1.3 A resolution and the search for differences between this protein and the homologous apoproteins A1 and C1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1S2Q
 
 | | Crystal structure of MAOB in complex with N-propargyl-1(R)-aminoindan (Rasagiline) | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-30 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
1S2R
 
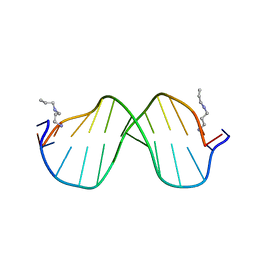 | | A High Resolution Crystal Structure of [d(CGCAAATTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3', SPERMINE | | Authors: | Woods, K.K, Maehigashi, T, Howerton, S.B, Tannenbaum, S, Williams, L.D. | | Deposit date: | 2004-01-09 | | Release date: | 2005-01-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of an extended A-tract: [d(CGCAAATTTGCG)]2.
J.Am.Chem.Soc., 126, 2004
|
|
1S2T
 
 | | Crystal Structure Of Apo Phosphoenolpyruvate Mutase | | Descriptor: | Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2U
 
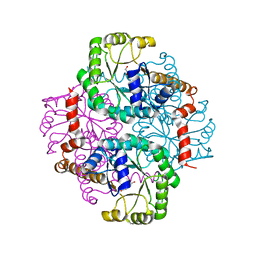 | | Crystal structure of the D58A phosphoenolpyruvate mutase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2V
 
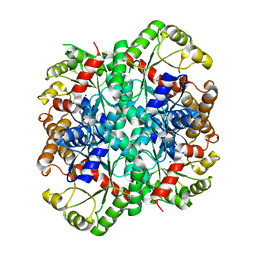 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | Descriptor: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2W
 
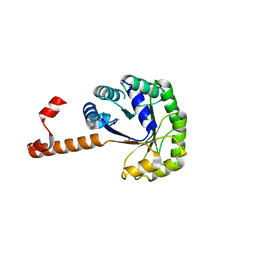 | | Crystal structure of phosphoenolpyruvate mutase in high ionic strength | | Descriptor: | Phosphoenolpyruvate phosphomutase, SULFATE ION | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2X
 
 | | Crystal structure of Cag-Z from Helicobacter pylori | | Descriptor: | Cag-Z, ISOPROPYL ALCOHOL | | Authors: | Cendron, L, Seydel, A, Angelini, A, Battistutta, R, Zanotti, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of CagZ, a protein from the Helicobacter pylori pathogenicity island that encodes for a type IV secretion system
J.Mol.Biol., 340, 2004
|
|
1S2Y
 
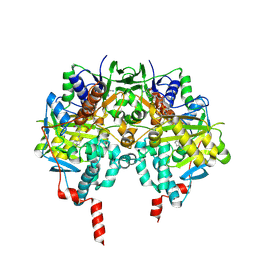 | | Crystal structure of MAOB in complex with N-propargyl-1(S)-aminoindan | | Descriptor: | Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-12 | | Release date: | 2004-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
1S2Z
 
 | | X-ray crystal structure of Desulfovibrio vulgaris Rubrerythrin with displacement of iron by zinc at the diiron Site | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.-J, Rose, J, Wang, B.-C. | | Deposit date: | 2004-01-12 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Displacement of iron by zinc at the diiron site of Desulfovibrio vulgaris rubrerythrin: X-ray crystal structure and anomalous scattering analysis
J.Inorg.Biochem., 98, 2004
|
|
1S30
 
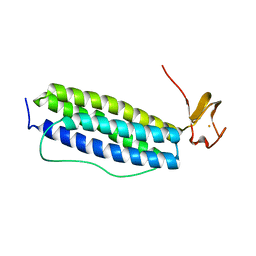 | | X-ray crystal structure of Desulfovibrio vulgaris Rubrerythrin with displacement of iron by zinc at the diiron Site | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.-J, Rose, J, Wang, B.-C. | | Deposit date: | 2004-01-12 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Displacement of iron by zinc at the diiron site of Desulfovibrio vulgaris rubrerythrin: X-ray crystal structure and anomalous scattering analysis
J.Inorg.Biochem., 98, 2004
|
|
1S31
 
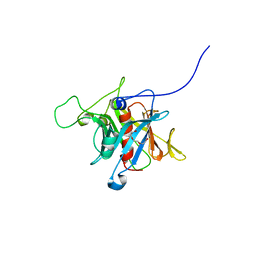 | | Crystal Structure Analysis of the human Tub protein (isoform a) spanning residues 289 through 561 | | Descriptor: | TRIETHYLENE GLYCOL, tubby isoform a | | Authors: | Boutboul, S, Carroll, K.J, Basdevant, A, Gomez, C, Nandrot, E, Clement, K, Shapiro, L, Abitbol, M. | | Deposit date: | 2004-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | A novel human obesity and sensory deficit syndrome resulting from a mutation in the TUB gene
To be Published
|
|
1S32
 
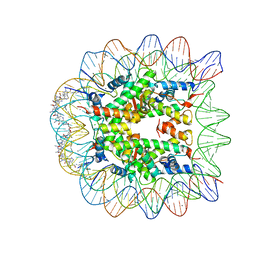 | | Molecular Recognition of the Nucleosomal 'Supergroove' | | Descriptor: | 2-(2-CARBAMOYLMETHOXY-ETHOXY)-ACETAMIDE, 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, ... | | Authors: | Edayathumangalam, R.S, Weyermann, P, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2004-01-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Recognition of the Nucleosomal 'Supergroove'
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S35
 
 | |
