7LXK
 
 | |
7LZL
 
 | |
7M0G
 
 | |
7M1D
 
 | |
7M1E
 
 | |
7M1W
 
 | |
7M25
 
 | |
7M27
 
 | |
7M28
 
 | |
7M29
 
 | |
7M2A
 
 | |
7M2B
 
 | |
7M2C
 
 | |
7M2M
 
 | | NMR Structure of GCAP5 | | Descriptor: | Guanylate cyclase activator 1A, MAGNESIUM ION | | Authors: | Ames, J.B, Cudia, D.L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR and EPR-DEER Structure of a Dimeric Guanylate Cyclase Activator Protein-5 from Zebrafish Photoreceptors.
Biochemistry, 60, 2021
|
|
7M3U
 
 | |
7M5T
 
 | |
7M67
 
 | |
7M73
 
 | |
7M77
 
 | |
7M79
 
 | |
7M7X
 
 | |
7MC1
 
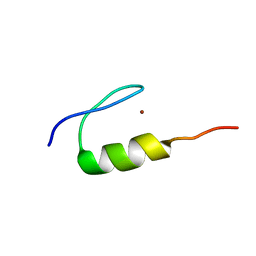 | | Solution structure of Miz-1 Zinc finger 10 | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7MC2
 
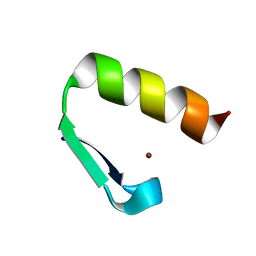 | | Solution structure of Miz-1 Zinc finger 11 H586Y | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7MC3
 
 | | Solution structure of Miz-1 zinc finger 12 | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7MJ3
 
 | | Structure of Hact-4 | | Descriptor: | Hact-4 | | Authors: | Schmidt, C.A, Daly, N.L. | | Deposit date: | 2021-04-19 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Diversity of coral derived peptides
To Be Published
|
|
