1TQ1
 
 | | Solution structure of At5g66040, a putative protein from Arabidosis Thaliana | | Descriptor: | senescence-associated family protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Singh, S, Lee, M.S, Tyler, E.M, Shahan, M.N, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a single-domain thiosulfate sulfurtransferase from Arabidopsis thaliana.
Protein Sci., 15, 2006
|
|
1TQR
 
 | | NMR Structure of DNA 17-mer GGAAAATCTCTAGCAGT corresponding to the extremity of the U5 LTR of the HIV-1 genome | | Descriptor: | DNA HIV-1 U5 LTR extremity | | Authors: | Renisio, J.G, Cosquer, S, Cherrak, I, El Antri, S, Mauffret, O, Fermandjian, S. | | Deposit date: | 2004-06-18 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Pre-organized structure of viral DNA at the binding-processing site of HIV-1 integrase
NUCLEIC ACIDS RES., 33, 2005
|
|
1TQZ
 
 | |
1TR4
 
 | | Solution structure of human oncogenic protein gankyrin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10 | | Authors: | Yuan, C, Li, J, Mahajan, A, Poi, M.J, Byeon, I.J, Tsai, M.D. | | Deposit date: | 2004-06-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human oncogenic protein gankyrin containing seven ankyrin repeats and analysis of its structure--function relationship.
Biochemistry, 43, 2004
|
|
1TR6
 
 | | NMR solution structure of omega-conotoxin [K10]GVIA, a cyclic cysteine knot peptide | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-13 | | Last modified: | 2011-10-05 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1TRL
 
 | | NMR SOLUTION STRUCTURE OF THE C-TERMINAL FRAGMENT 255-316 OF THERMOLYSIN: A DIMER FORMED BY SUBUNITS HAVING THE NATIVE STRUCTURE | | Descriptor: | THERMOLYSIN FRAGMENT 255 - 316 | | Authors: | Rico, M, Jimenez, M.A, Gonzalez, C, De Filippis, V, Fontana, A. | | Deposit date: | 1994-09-02 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the C-terminal fragment 255-316 of thermolysin: a dimer formed by subunits having the native structure.
Biochemistry, 33, 1994
|
|
1TT3
 
 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TTD
 
 | | SOLUTION-STATE STRUCTURE OF A DNA DODECAMER DUPLEX CONTAINING A CIS-SYN THYMINE CYCLOBUTANE DIMER | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*AP*TP*TP*CP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*GP*AP*AP*(TTD)P*AP*AP*G)-3') | | Authors: | Mcateer, K, Jing, Y, Kao, J, Taylor, J.-S, Kennedy, M.A. | | Deposit date: | 1999-01-20 | | Release date: | 1999-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA.
J.Mol.Biol., 282, 1998
|
|
1TTE
 
 | |
1TTL
 
 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1TTN
 
 | |
1TTV
 
 | | NMR Structure of a Complex Between MDM2 and a Small Molecule Inhibitor | | Descriptor: | 1-{[4,5-BIS(4-CHLOROPHENYL)-2-(2-ISOPROPOXY-4-METHOXYPHENYL)-4,5-DIHYDRO-1H-IMIDAZOL-1-YL]CARBONYL}PIPERAZINE, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Fry, D.C, Emerson, S.D, Palme, S, Vu, B.T, Liu, C.M, Podlaski, F. | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex between MDM2 and a small molecule inhibitor.
J.Biomol.Nmr, 30, 2004
|
|
1TTX
 
 | | Solution Structure of human beta parvalbumin (oncomodulin) refined with a paramagnetism based strategy | | Descriptor: | CALCIUM ION, Oncomodulin | | Authors: | Babini, E, Bertini, I, Capozzi, F, Del Bianco, C, Hollender, D, Kiss, T, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human beta-Parvalbumin and Structural Comparison with Its Paralog alpha-Parvalbumin and with Their Rat Orthologs(,)
Biochemistry, 43, 2004
|
|
1TUJ
 
 | | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate | | Descriptor: | 3-TRIMETHYLSILYL-PROPIONATE-2,2,3,3,-D4, odorant binding protein ASP2 | | Authors: | Lescop, E, Briand, L, Pernollet, J.-C, Guittet, E. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate
To be Published
|
|
1TUM
 
 | | MUTT PYROPHOSPHOHYDROLASE-METAL-NUCLEOTIDE-METAL COMPLEX, NMR, 16 STRUCTURES | | Descriptor: | COBALT TETRAAMMINE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lin, J, Abeygunawardana, C, Frick, D.N, Bessman, M.J, Mildvan, A.S. | | Deposit date: | 1996-12-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the quaternary MutT-M2+-AMPCPP-M2+ complex and mechanism of its pyrophosphohydrolase action.
Biochemistry, 36, 1997
|
|
1TUQ
 
 | | NMR Structure Analysis of the B-DNA Dodecamer CTCtCACGTGGAG with a tricyclic cytosin base analogue | | Descriptor: | 5'-D(P*CP*TP*CP*(TC1)P*AP*CP*GP*TP*GP*GP*AP*G)-3' | | Authors: | Engman, K.C, Sandin, P, Osborne, S, Brown, T, Billeter, M, Lincoln, P, Norden, B, Albinsson, B, Wilhelmsson, L.M. | | Deposit date: | 2004-06-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA adopts normal B-form upon incorporation of highly fluorescent DNA base analogue tC: NMR structure and UV-Vis spectroscopy characterization.
Nucleic Acids Res., 32, 2004
|
|
1TUR
 
 | | SOLUTION STRUCTURE OF TURKEY OVOMUCOID THIRD DOMAIN AS DETERMINED FROM NUCLEAR MAGNETIC RESONANCE DATA | | Descriptor: | OVOMUCOID | | Authors: | Krezel, A.M, Darba, P, Robertson, A.D, Fejzo, J, Macura, S, Markley, J.L. | | Deposit date: | 1994-07-06 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of turkey ovomucoid third domain as determined from nuclear magnetic resonance data.
J.Mol.Biol., 242, 1994
|
|
1TUS
 
 | |
1TUT
 
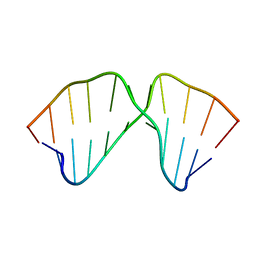 | | J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns | | Descriptor: | 5'-R(*GP*AP*GP*GP*AP*AP*GP*GP*CP*GP*A)-3', 5'-R(*UP*CP*GP*UP*UP*AP*AP*UP*CP*UP*C)-3' | | Authors: | Znosko, B.M, Kennedy, S.D, Wille, P.C, Krugh, T.R, Turner, D.H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Features and Thermodynamics of the J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns.
Biochemistry, 43, 2004
|
|
1TUZ
 
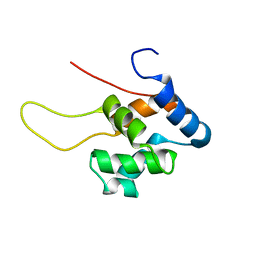 | | NMR Structure of the Diacylglycerol kinase alpha, NESGC target HR532 | | Descriptor: | diacylglycerol kinase alpha | | Authors: | Liu, G, Shao, Y, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Diacylglycerol kinase alpha, NESGC target HR532
To be Published
|
|
1TV0
 
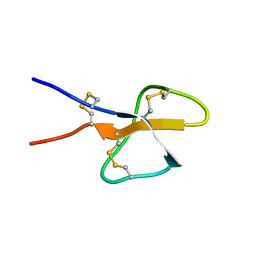 | | Solution structure of cryptdin-4, the most potent alpha-defensin from mouse Paneth cells | | Descriptor: | Cryptdin-4 | | Authors: | Jing, W, Hunter, H.N, Tanabe, H, Ouellette, A.J, Vogel, H.J. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cryptdin-4, a Mouse Paneth Cell alpha-Defensin.
Biochemistry, 43, 2004
|
|
1TVC
 
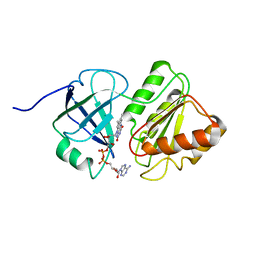 | | FAD and NADH binding domain of methane monooxygenase reductase from Methylococcus capsulatus (Bath) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, METHANE MONOOXYGENASE COMPONENT C | | Authors: | Chatwood, L.L, Mueller, J, Gross, J.D, Wagner, G, Lippard, S.J. | | Deposit date: | 2004-06-29 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Flavin Domain from Soluble Methane Monooxygenase Reductase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
1TVI
 
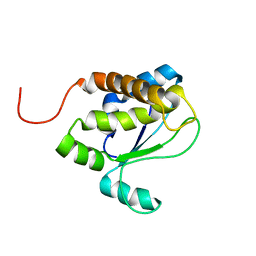 | | Solution structure of TM1509 from Thermotoga maritima: VT1, a NESGC target protein | | Descriptor: | Hypothetical UPF0054 protein TM1509 | | Authors: | Penhoat, C.H, Atreya, H.S, Kim, S, Li, Z, Yee, A, Xiao, R, Murray, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1TVJ
 
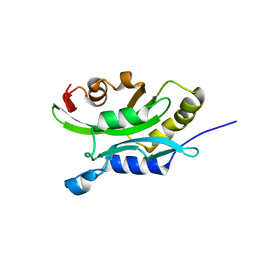 | | Solution Structure of chick cofilin | | Descriptor: | Cofilin | | Authors: | Gorbatyuk, V.Y, Nosworthy, N.J, Robson, S.A, Maciejewski, M.W, dos Remedios, C.G, King, G.F. | | Deposit date: | 2004-06-29 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Study of the Molecular Basis for Phosphoinositide Regulation of the Cofilin-Actin Interactions.
To be Published
|
|
1TVM
 
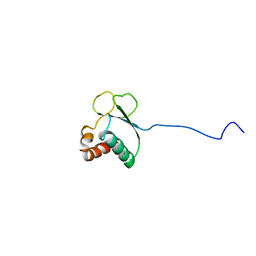 | | NMR structure of enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system | | Descriptor: | PTS system, galactitol-specific IIB component | | Authors: | Volpon, L, Young, C.R, Lim, N.S, Iannuzzi, P, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system and its interaction with GatA.
Protein Sci., 15, 2006
|
|
