6U19
 
 | |
6U1L
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
6U1O
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
6U24
 
 | | NMR solution structure of triazole bridged SFTI-1 | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U3R
 
 | |
6U3S
 
 | |
6U46
 
 | |
6U4M
 
 | | Solution structure of paxillin LIM4 | | Descriptor: | Paxillin, ZINC ION | | Authors: | Zhu, L, Qin, J. | | Deposit date: | 2019-08-26 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Paxillin Recruitment by Kindlin-2 in Regulating Cell Adhesion.
Structure, 27, 2019
|
|
6U4N
 
 | |
6U6G
 
 | |
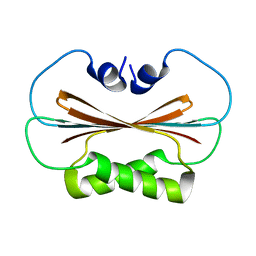
6U6P
 
 | |
6U6Q
 
 | |
6U6R
 
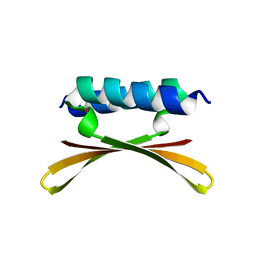 | |
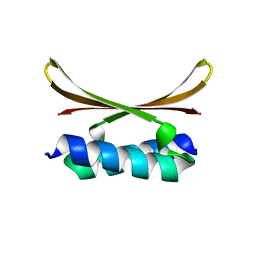
6U6S
 
 | |
6U79
 
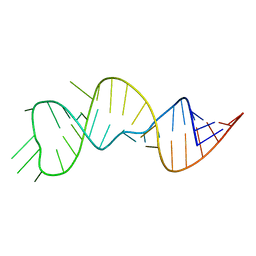 | |
6U7Q
 
 | | NMR solution structure of SFTI-R10 | | Descriptor: | GLY-ARG-CYS-THR-LYS-SER-ILE-PRO-PRO-ARG-CYS-PHE-PRO-ASP inhibitor | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7R
 
 | |
6U7S
 
 | |
6U7U
 
 | | NMR solution structure of triazole bridged matriptase inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ARG-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7W
 
 | | NMR solution structure of a triazole bridged KLK7 inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-LYS-ALA-LEU-PHE-SER-ASN-PRO-PRO-ILE-ALA-PHE-PRO-ASN | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7X
 
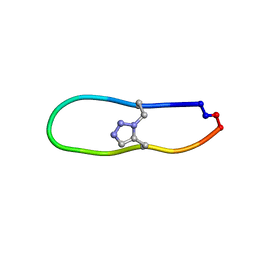 | | NMR solution structure of triazole bridged plasmin inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Wang, C.K, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UCH
 
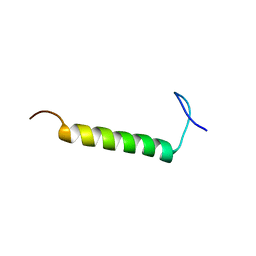 | | SMARCB1 nucleosome-interacting C-terminal alpha helix | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Valencia, A.M, Sun, Z.Y.J, Seo, H.S, Vangos, H.S, Yeoh, Z.C, Mashtalir, N, Dhe-Paganon, S, Kadoch, C. | | Deposit date: | 2019-09-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling.
Cell, 179, 2019
|
|
6UCJ
 
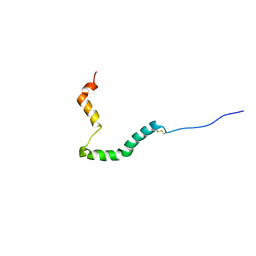 | | proIAPP in DPC Micelles - Two-Conformer Ensemble Refinement, Open Conformer | | Descriptor: | Islet amyloid polypeptide | | Authors: | DeLisle, C.F, Malooley, A.L, Banerjee, I, Lorieau, J.L. | | Deposit date: | 2019-09-16 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment.
Febs J., 287, 2020
|
|
6UCK
 
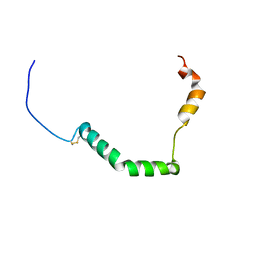 | | proIAPP in DPC Micelles - Two-Conformer Ensemble Refinement, Bent Conformer | | Descriptor: | Islet amyloid polypeptide | | Authors: | DeLisle, C.F, Malooley, A.L, Banerjee, I, Lorieau, J.L. | | Deposit date: | 2019-09-16 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment.
Febs J., 287, 2020
|
|
6UCO
 
 | |
