1HY8
 
 | | SOLUTION STRUCTURE OF B. SUBTILIS ACYL CARRIER PROTEIN | | Descriptor: | ACYL CARRIER PROTEIN | | Authors: | Xu, G.-Y, Tam, A, Lin, L, Hixon, J, Fritz, C.C, Power, R. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of B. subtilis acyl carrier protein.
Structure, 9, 2001
|
|
1HZ0
 
 | | NMR STRUCTURE OF THE 2-AMINO-1-METHYL-6-PHENYLIMIDAZO[4,5-B]PYRIDINE (PHIP) C8-DEOXYGUANOSINE ADDUCT IN DUPLEX DNA | | Descriptor: | 2-AMINO-1-METHYL-6-PHENYLIMIDAZO[4,5-B]PYRIDINE, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Brown, K, Cosman, M. | | Deposit date: | 2001-01-23 | | Release date: | 2001-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 2-amino-1- methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HZ2
 
 | | SOLUTION NMR STRUCTURE OF SELF-COMPLEMENTARY DUPLEX 5'-D(AGGCG*CCT)2 CONTAINING A TRIMETHYLENE CROSSLINK AT THE N2 POSITION OF G*. MODEL OF A MALONDIALDEHYDE CROSSLINK | | Descriptor: | DNA (5'-D(*AP*GP*GP*CP*GP*CP*CP*T)-3'), PROPANE | | Authors: | Dooley, P.A, Tsarouhtsis, D, Korbel, G.A, Nechev, L.V, Shearer, J, Zegar, I.S, Harris, C.M, Stone, M.P, Harris, T.M. | | Deposit date: | 2001-01-23 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of an oligodeoxynucleotide containing a trimethylene interstrand cross-link in a 5'-(CpG) motif: model of a malondialdehyde cross-link.
J.Am.Chem.Soc., 123, 2001
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | Descriptor: | A-BETA AMYLOID | | Authors: | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
1HZ8
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A CONCATEMER OF EGF-HOMOLOGY MODULES OF THE HUMAN LOW DENSITY LIPOPROTEIN RECEPTOR | | Descriptor: | CALCIUM ION, LOW DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Aliabadizadeh, K, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2001-01-23 | | Release date: | 2001-08-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure and backbone dynamics of a concatemer of epidermal growth factor homology modules of the human low-density lipoprotein receptor.
J.Mol.Biol., 311, 2001
|
|
1HZE
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-24 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
1HZK
 
 | |
1HZL
 
 | | SOLUTION STRUCTURES OF C-1027 APOPROTEIN AND ITS COMPLEX WITH THE AROMATIZED CHROMOPHORE | | Descriptor: | C-1027 APOPROTEIN, C-1027 AROMATIZED CHROMOPHORE | | Authors: | Tanaka, T, Fukuda-Ishisaka, S, Hirama, M, Otani, T. | | Deposit date: | 2001-01-25 | | Release date: | 2001-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of C-1027 apoprotein and its complex with the aromatized chromophore.
J.Mol.Biol., 309, 2001
|
|
1I0U
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A CONCATEMER OF EGF-HOMOLOGY MODULES OF THE HUMAN LOW DENSITY LIPOPROTEIN RECEPTOR | | Descriptor: | CALCIUM ION, LOW DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Aliabadizadeh, K, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2001-01-29 | | Release date: | 2001-08-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure and backbone dynamics of a concatemer of epidermal growth factor homology modules of the human low-density lipoprotein receptor.
J.Mol.Biol., 311, 2001
|
|
1I11
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN, SOX-5 HMG BOX FROM MOUSE | | Descriptor: | TRANSCRIPTION FACTOR SOX-5 | | Authors: | Cary, P.D, Read, C.M, Davis, B, Driscoll, P.C, Crane-Robinson, C. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the DNA-binding domain of mouse Sox-5.
Protein Sci., 10, 2001
|
|
1I17
 
 | | NMR STRUCTURE OF MOUSE DOPPEL 51-157 | | Descriptor: | PRION-LIKE PROTEIN | | Authors: | Mo, H, Moore, R.C, Cohen, F.E, Westaway, D, Prusiner, S.B, Wright, P.E, Dyson, H.J. | | Deposit date: | 2001-01-31 | | Release date: | 2001-03-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two different neurodegenerative diseases caused by proteins with similar structures.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1I18
 
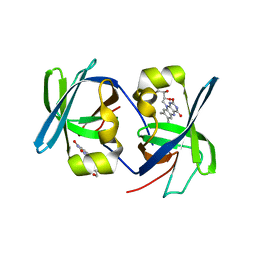 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-31 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
1I25
 
 | |
1I2U
 
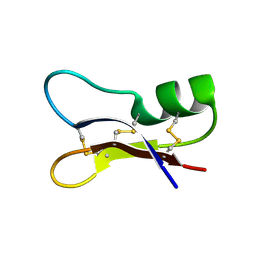 | | NMR SOLUTION STRUCTURES OF ANTIFUNGAL HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
1I2V
 
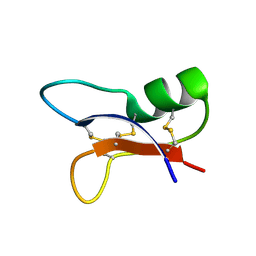 | | NMR SOLUTION STRUCTURES OF AN ANTIFUNGAL AND ANTIBACTERIAL MUTANT OF HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
1I3X
 
 | |
1I3Y
 
 | |
1I42
 
 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 | | Descriptor: | P47 | | Authors: | Yuan, X, Shaw, A, Zhang, X, Kondo, H, Lally, J, Freemont, P.S, Matthews, S. | | Deposit date: | 2001-02-19 | | Release date: | 2001-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
1I46
 
 | |
1I4B
 
 | |
1I4C
 
 | |
1I5H
 
 | |
1I5J
 
 | |
1I5T
 
 | | SOLUTION STRUCTURE OF CYANOFERRICYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Yao, Y, Qian, C, Ye, K, Wang, J, Tang, W. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cyanoferricytochrome c: ligand-controlled conformational flexibility and electronic structure of the heme moiety.
J.Biol.Inorg.Chem., 7, 2002
|
|
1I5U
 
 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
