1FYC
 
 | | INNER LIPOYL DOMAIN FROM HUMAN PYRUVATE DEHYDROGENASE (PDH) COMPLEX, NMR, 1 STRUCTURE | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE (E2P) | | Authors: | Howard, M.J, Fuller, C, Broadhurst, R.W, Quinn, J, Yeaman, S.J, Perham, R.N. | | Deposit date: | 1997-02-21 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the major autoantigen in primary biliary cirrhosis.
Gastroenterology, 115, 1998
|
|
1FYI
 
 | |
1FYJ
 
 | | SOLUTION STRUCTURE OF MULTI-FUNCTIONAL PEPTIDE MOTIF-1 PRESENT IN HUMAN GLUTAMYL-PROLYL TRNA SYNTHETASE (EPRS). | | Descriptor: | MULTIFUNCTIONAL AMINOACYL-TRNA SYNTHETASE | | Authors: | Jeong, E.-J, Hwang, G.-S, Kim, K.H, Kim, M.J, Kim, S, Kim, K.-S. | | Deposit date: | 2000-10-02 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of multifunctional peptide motifs in human bifunctional tRNA synthetase: identification of RNA-binding residues and functional implications for tandem repeats.
Biochemistry, 39, 2000
|
|
1FZL
 
 | | DNA WITH PYRENE PAIRED AT ABASIC SITES | | Descriptor: | 5'-D(*CP*AP*CP*AP*AP*AP*CP*AP*(PYP)P*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*(3DR)P*TP*GP*TP*TP*TP*GP*TP*G)-3' | | Authors: | Smirnov, S, Matray, T.J, Kool, E.T, de los Santos, C. | | Deposit date: | 2000-10-03 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Integrity of duplex structures without hydrogen bonding: DNA with pyrene paired at abasic sites
Nucleic Acids Res., 30, 2002
|
|
1FZS
 
 | | DNA WITH PYRENE PAIRED AT ABASIC SITE | | Descriptor: | 5'-D(*CP*AP*CP*AP*AP*AP*CP*AP*(PYP))-3', 5'-D(*GP*TP*GP*CP*(3DR)P*TP*GP*TP*TP*TP*GP*TP*G)-3' | | Authors: | Smirnov, S, Matray, T.J, Kool, E.T, de los Santos, C. | | Deposit date: | 2000-10-04 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Integrity of duplex structures without hydrogen bonding: DNA with pyrene paired at abasic sites
Nucleic Acids Res., 30, 2002
|
|
1FZT
 
 | | SOLUTION STRUCTURE AND DYNAMICS OF AN OPEN B-SHEET, GLYCOLYTIC ENZYME-MONOMERIC 23.7 KDA PHOSPHOGLYCERATE MUTASE FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PHOSPHOGLYCERATE MUTASE | | Authors: | Uhrinova, S, Uhrin, D, Nairn, J, Price, N.C, Fothergill-Gilmore, L.A. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of an open beta-sheet, glycolytic enzyme, monomeric 23.7 kDa phosphoglycerate mutase from Schizosaccharomyces pombe.
J.Mol.Biol., 306, 2001
|
|
1FZX
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAAAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
1G03
 
 | | NMR STRUCTURE OF N-TERMINAL DOMAIN OF HTLV-I CA1-134 | | Descriptor: | HTLV-I CAPSID PROTEIN | | Authors: | Cornilescu, C.C, Bouamr, F, Yao, X, Carter, C, Tjandra, N. | | Deposit date: | 2000-10-05 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein.
J.Mol.Biol., 306, 2001
|
|
1G04
 
 | | SOLUTION STRUCTURE OF SYNTHETIC 26-MER PEPTIDE CONTAINING 145-169 SHEEP PRION PROTEIN SEGMENT AND C-TERMINAL CYSTEINE | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Kozin, S.A, Bertho, G, Mazur, A.K, Rabesona, H, Girault, J.-P, Haertle, T, Takahashi, M, Debey, P, Hui Bon Hoa, G. | | Deposit date: | 2000-10-05 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sheep prion protein synthetic peptide spanning helix 1 and beta-strand 2 (residues 142-166) shows beta-hairpin structure in solution.
J.Biol.Chem., 276, 2001
|
|
1G10
 
 | | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR PROTEIN NMR STRUCTURE | | Descriptor: | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR | | Authors: | Hemmi, H, Studts, J.M, Chae, Y.K, Song, J, Markley, J.L, Fox, B.G. | | Deposit date: | 2000-10-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the toluene 4-monooxygenase effector protein (T4moD).
Biochemistry, 40, 2001
|
|
1G11
 
 | | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR PROTEIN NMR STRUCTURE | | Descriptor: | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR | | Authors: | Hemmi, H, Studts, J.M, Chae, Y.K, Song, J, Markley, J.L, Fox, B.G. | | Deposit date: | 2000-10-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the toluene 4-monooxygenase effector protein (T4moD).
Biochemistry, 40, 2001
|
|
1G14
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAGAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*CP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*GP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
1G1E
 
 | | NMR STRUCTURE OF THE HUMAN MAD1 TRANSREPRESSION DOMAIN SID IN COMPLEX WITH MAMMALIAN SIN3A PAH2 DOMAIN | | Descriptor: | MAD1 PROTEIN, SIN3A | | Authors: | Brubaker, K, Cowley, S.M, Huang, K, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G1N
 
 | | NICKED DECAMER DNA WITH PEG6 TETHER, NMR, 30 STRUCTURES | | Descriptor: | 5'-D(*GP*TP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*AP*CP*AP*AP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*TP*T)-3', ... | | Authors: | Bocian, W, Kozerski, L, Mazurek, A.P, Kawecki, R. | | Deposit date: | 2000-10-13 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A nicked duplex decamer DNA with a PEG(6) tether.
Nucleic Acids Res., 29, 2001
|
|
1G1P
 
 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Barbier, J, Gilles, N, Molgo, J, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-13 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
1G1Z
 
 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels, LEU12-PRO13 Cis isomer | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Le Gall, F, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-16 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
1G22
 
 | |
1G25
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE HUMAN TFIIH MAT1 SUBUNIT | | Descriptor: | CDK-ACTIVATING KINASE ASSEMBLY FACTOR MAT1, ZINC ION | | Authors: | Gervais, V, Wasielewski, E, Busso, D, Poterszman, A, Egly, J.M, Thierry, J.C, Kieffer, B. | | Deposit date: | 2000-10-17 | | Release date: | 2000-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Domain of the Human TFIIH MAT1 Subunit: New Insights into the RING Finger Family
J.Biol.Chem., 276, 2001
|
|
1G26
 
 | |
1G2G
 
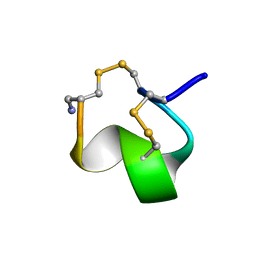 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | Descriptor: | ALPHA-CONOTOXIN IMI | | Authors: | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
1G2S
 
 | |
1G2T
 
 | |
1G3A
 
 | |
1G47
 
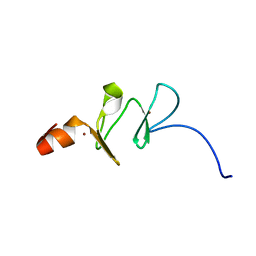 | | 1ST LIM DOMAIN OF PINCH PROTEIN | | Descriptor: | PINCH PROTEIN, ZINC ION | | Authors: | Velyvis, A, Yang, Y, Wu, C, Qin, J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the focal adhesion adaptor PINCH LIM1 domain and characterization of its interaction with the integrin-linked kinase ankyrin repeat domain.
J.Biol.Chem., 276, 2001
|
|
1G4F
 
 | | NMR STRUCTURE OF THE FIFTH DOMAIN OF HUMAN BETA2-GLYCOPROTEIN I | | Descriptor: | BETA2-GLYCOPROTEIN I | | Authors: | Hoshino, M, Hagihara, Y, Nishii, I, Yamazaki, T, Kato, H, Goto, Y. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid-binding site of human beta(2)-glycoprotein I domain V by heteronuclear magnetic resonance.
J.Mol.Biol., 304, 2000
|
|
