1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | Descriptor: | orf, hypothetical protein | | Authors: | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
1P9Z
 
 | | The Solution Structure of Antifungal Peptide Distinct With a Five-disulfide Motif from Eucommia ulmoides Oliver | | Descriptor: | Eucommia Antifungal peptide 2 | | Authors: | Huang, R.H, Xiang, Y, Tu, G.Z, Zhang, Y, Wang, D.C. | | Deposit date: | 2003-05-13 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Eucommia Antifungal Peptide: A Novel Structural Model Distinct with a Five-Disulfide Motif.
Biochemistry, 43, 2004
|
|
1PA4
 
 | | Solution structure of a putative ribosome-binding factor from Mycoplasma pneumoniae (MPN156) | | Descriptor: | Probable ribosome-binding factor A | | Authors: | Rubin, S.M, Pelton, J.G, Yokota, H, Kim, R, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-05-13 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative ribosome binding protein from Mycoplasma pneumoniae and comparison to a distant homolog.
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1PAN
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAO PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1PAV
 
 | | SOLUTION NMR STRUCTURE OF HYPOTHETICAL PROTEIN TA1414 OF THERMOPLASMA ACIDOPHILUM | | Descriptor: | Hypothetical protein Ta1170/Ta1414 | | Authors: | Monleon, D, Yee, A, Liu, C.S, Arrowsmith, C, Celda, B. | | Deposit date: | 2003-05-14 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein TA1414 from Thermoplasma acidophilum.
J.Biomol.Nmr, 28, 2004
|
|
1PB5
 
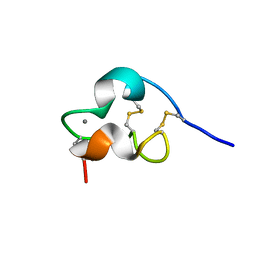 | | NMR Structure of a Prototype LNR Module from Human Notch1 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 1 | | Authors: | Vardar, D, North, C.L, Sanchez-Irizarry, C, Aster, J.C, Blacklow, S.C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of a Prototype Lin12-Notch Repeat Module from Human Notch1
Biochemistry, 42, 2003
|
|
1PBA
 
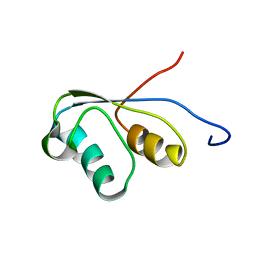 | | THE NMR STRUCTURE OF THE ACTIVATION DOMAIN ISOLATED FROM PORCINE PROCARBOXYPEPTIDASE B | | Descriptor: | PROCARBOXYPEPTIDASE B | | Authors: | Vendrell, J, Wider, G, Billeter, M, Aviles, F.X, Wuthrich, K. | | Deposit date: | 1991-11-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the activation domain isolated from porcine procarboxypeptidase B.
EMBO J., 10, 1991
|
|
1PBU
 
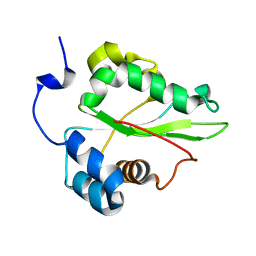 | | Solution structure of the C-terminal domain of the human eEF1Bgamma subunit | | Descriptor: | Elongation factor 1-gamma | | Authors: | Vanwetswinkel, S, Kriek, J, Andersen, G.R, Guntert, P, Dijk, J, Canters, G.W, Siegal, G. | | Deposit date: | 2003-05-15 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, (15)N and (13)C resonance assignments of the highly conserved 19 kDa C-terminal domain from human Elongation Factor 1Bgamma.
J.Biomol.Nmr, 26, 2003
|
|
1PBZ
 
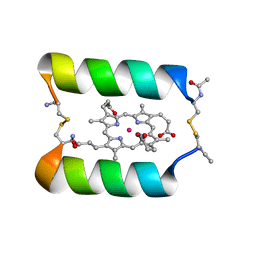 | |
1PC0
 
 | |
1PC2
 
 | |
1PCE
 
 | | SOLUTION STRUCTURE AND DYNAMICS OF PEC-60, A PROTEIN OF THE KAZAL TYPE INHIBITOR FAMILY, DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | PEC-60 | | Authors: | Liepinsh, E, Berndt, K.D, Sillard, R, Mutt, V, Otting, G. | | Deposit date: | 1994-02-22 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of PEC-60, a protein of the Kazal type inhibitor family, determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 239, 1994
|
|
1PD6
 
 | |
1PD7
 
 | | Extended SID of Mad1 bound to the PAH2 domain of mSin3B | | Descriptor: | Mad1, Sin3b protein | | Authors: | Van Ingen, H, Lasonder, E, Jansen, J.F, Kaan, A.M, Spronk, C.A, Stunnenberg, H.G, Vuister, G.W. | | Deposit date: | 2003-05-19 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Extension of the binding motif of the sin3 interacting domain of the mad family proteins(,).
Biochemistry, 43, 2004
|
|
1PDC
 
 | |
1PDT
 
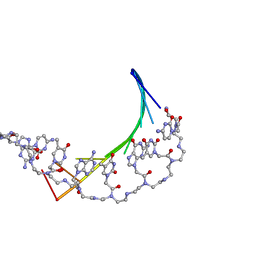 | | PD235, PNA-DNA DUPLEX, NMR, 8 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*AP*GP*C)-3', PEPTIDE NUCLEIC ACID (COOH-P(*G*C*T*A*T*G*T*C)-NH2) | | Authors: | Eriksson, M, Nielsen, P.E. | | Deposit date: | 1996-03-28 | | Release date: | 1996-10-14 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a peptide nucleic acid-DNA duplex.
Nat.Struct.Biol., 3, 1996
|
|
1PDX
 
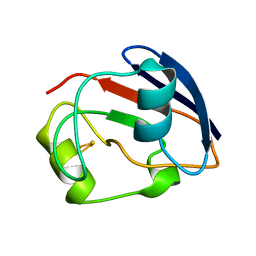 | | PUTIDAREDOXIN | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (PUTIDAREDOXIN) | | Authors: | Pochapsky, T.C, Jain, N.U, Kuti, M, Lyons, T.A, Heymont, J. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A refined model for the solution structure of oxidized putidaredoxin.
Biochemistry, 38, 1999
|
|
1PE3
 
 | |
1PEH
 
 | | NMR STRUCTURE OF THE MEMBRANE-BINDING DOMAIN OF CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE, 10 STRUCTURES | | Descriptor: | PEPNH1 | | Authors: | Dunne, S.J, Cornell, R.B, Johnson, J.E, Glover, N.R, Tracey, A.S. | | Deposit date: | 1996-06-10 | | Release date: | 1996-12-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase.
Biochemistry, 35, 1996
|
|
1PEI
 
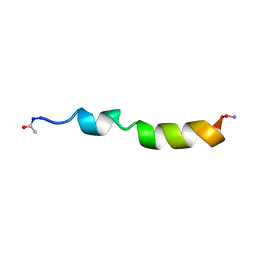 | | NMR STRUCTURE OF THE MEMBRANE-BINDING DOMAIN OF CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE, 10 STRUCTURES | | Descriptor: | PEPC22 | | Authors: | Dunne, S.J, Cornell, R.B, Johnson, J.E, Glover, N.R, Tracey, A.S. | | Deposit date: | 1996-06-10 | | Release date: | 1996-12-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase.
Biochemistry, 35, 1996
|
|
1PET
 
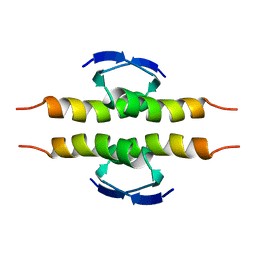 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
1PFD
 
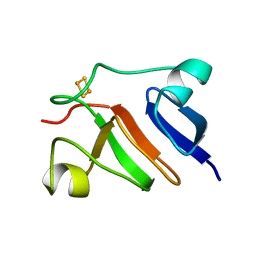 | | THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN, NMR, 18 STRUCTURES | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Im, S.-C, Liu, G, Luchinat, C, Sykes, A.G, Bertini, I. | | Deposit date: | 1998-05-05 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of parsley [2Fe-2S]ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
1PFJ
 
 | | Solution structure of the N-terminal PH/PTB domain of the TFIIH P62 subunit | | Descriptor: | TFIIH basal transcription factor complex p62 subunit | | Authors: | Gervais, V, Lamour, V, Jawhari, A, Frindel, F, Wasielewski, E, Thierry, J.C, Kieffer, B, Poterszman, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | TFIIH contains a PH domain involved in DNA nucleotide excision repair.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PFL
 
 | | REFINED SOLUTION STRUCTURE OF HUMAN PROFILIN I | | Descriptor: | PROFILIN I | | Authors: | Metzler, W.J, Farmer II, B.T, Constantine, K.L, Friedrichs, M.S, Lavoie, T, Mueller, L. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of human profilin I.
Protein Sci., 4, 1995
|
|
1PFM
 
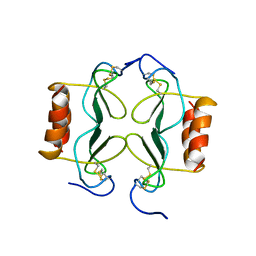 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 1-15 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
