6MF8
 
 | | TCR alpha transmembrane domain | | Descriptor: | T-cell receptor alpha chain C region | | Authors: | Brazin, K.N, Reinherz, E.L. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The T Cell Antigen Receptor alpha Transmembrane Domain Coordinates Triggering through Regulation of Bilayer Immersion and CD3 Subunit Associations.
Immunity, 49, 2018
|
|
6MG9
 
 | |
6MI5
 
 | | NMR solution structure of lanmodulin (LanM) complexed with yttrium(III) ions | | Descriptor: | Lanmodulin, YTTRIUM (III) ION | | Authors: | Cook, E.C, Featherson, E.R, Showalter, S.A, Cotruvo Jr, J.A. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Rare Earth Element Recognition by Methylobacterium extorquens Lanmodulin.
Biochemistry, 58, 2019
|
|
6MI9
 
 | |
6MIE
 
 | | Solution NMR structure of the KCNQ1 voltage-sensing domain | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Taylor, K.C, Kuenze, G, Smith, J.A, Meiler, J, McFeeters, R.L, Sanders, C.R. | | Deposit date: | 2018-09-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state.
Elife, 9, 2020
|
|
6MIF
 
 | | Lim5 domain of PINCH1 protein | | Descriptor: | LIM and senescent cell antigen-like-containing domain protein 1, ZINC ION | | Authors: | Qin, J, Vaynberg, J. | | Deposit date: | 2018-09-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Non-catalytic signaling by pseudokinase ILK for regulating cell adhesion.
Nat Commun, 9, 2018
|
|
6MJD
 
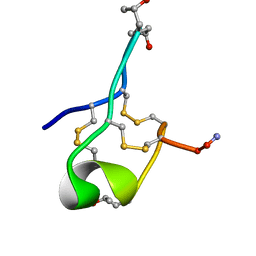 | | NMR Solution structure of GIIIC | | Descriptor: | ARG-ASP-CYS-CYS-THR-HYP-HYP-LYS-LYS-CYS-LYS-ASP-ARG-ARG-CYS-LYS-HYP-LEU-LYS-CYS-CYS-ALA-NH2 | | Authors: | Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2018-09-20 | | Release date: | 2018-11-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of mu-Conotoxin GIIIC: Leucine 18 Induces Local Repacking of the N-Terminus Resulting in Reduced NaVChannel Potency.
Molecules, 23, 2018
|
|
6MJV
 
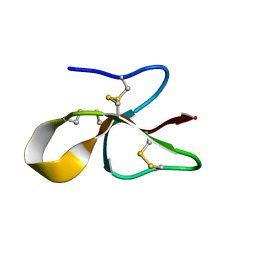 | | A consensus human beta defensin | | Descriptor: | Human beta-defensin | | Authors: | Amero, C, Villegas-Moreno, J, Rodriguez, A, Norton, R.S, Corzo, G. | | Deposit date: | 2018-09-22 | | Release date: | 2019-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial activity and structure of a consensus human beta-defensin and its comparison to a novel putative hBD10.
Proteins, 88, 2020
|
|
6MK4
 
 | |
6MK5
 
 | |
6MK7
 
 | | Solution structure of the large extracellular loop of FtsX in Streptococcus pneumoniae | | Descriptor: | Cell division protein FtsX | | Authors: | Edmonds, K.A, Fu, Y, Wu, H, Rued, B.E, Bruce, K.E, Winkler, M.E, Giedroc, D.P. | | Deposit date: | 2018-09-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
MBio, 10, 2019
|
|
6MK8
 
 | |
6MM4
 
 | |
6MNL
 
 | |
6MNT
 
 | |
6MPO
 
 | |
6MPP
 
 | |
6MSP
 
 | | De novo Designed Protein Foldit3 | | Descriptor: | De novo Designed Protein Foldit3 | | Authors: | Liu, G, Ishida, Y, Swapna, G.V.T, Kleinfelter, S, Koepnick, B, Baker, D, Montelione, G.T. | | Deposit date: | 2018-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MUN
 
 | | Structure of hRpn10 bound to UBQLN2 UBL | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquilin-2 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2018-10-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome.
J.Mol.Biol., 431, 2019
|
|
6MV3
 
 | | NMR structure of the cNTnC-cTnI chimera bound to calcium desensitizer W7 | | Descriptor: | CALCIUM ION, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE, Troponin C, ... | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2018-10-24 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Changes Induced by the Binding of the Calcium Desensitizer W7 to Cardiac Troponin.
Biochemistry, 57, 2018
|
|
6MW6
 
 | |
6MWM
 
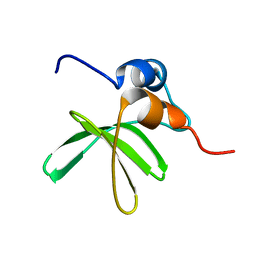 | | Bat coronavirus HKU4 SUD-C | | Descriptor: | Non-structural protein 3 | | Authors: | Staup, A.J, De Silva, I.U, Catt, J.T, Tan, X, Hammond, R.G, Johnson, M.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the SARS-Unique Domain C From the Bat Coronavirus HKU4.
Nat Prod Commun, 14, 2019
|
|
6MXQ
 
 | |
6MY1
 
 | | Solution structure of gomesin at 278 K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
6MY2
 
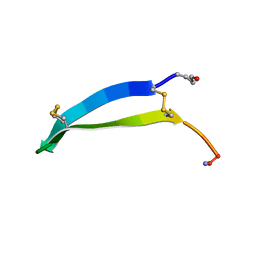 | | Solution structure of gomesin at 298 K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
