2N31
 
 | |
2N32
 
 | | NMR solution structure of the N-terminal domain of NisI, a lipoprotein from Lactococcus lactis which confers immunity against nisin | | Descriptor: | Nisin immunity protein | | Authors: | Hacker, C, Christ, N.A, Korn, S, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Entian, K, Woehnert, J. | | Deposit date: | 2015-05-21 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin.
J.Biol.Chem., 290, 2015
|
|
2N34
 
 | | NMR assignments and solution structure of the JAK interaction region of SOCS5 | | Descriptor: | Suppressor of cytokine signaling 5 | | Authors: | Chandrashekaran, I.R, Mohanty, B, Linossi, E.M, Nicholson, S.E, Babon, J, Norton, R.S, Dagley, L.F, Leung, E.W.W, Murphy, J.M. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5.
Biochemistry, 54, 2015
|
|
2N35
 
 | |
2N37
 
 | | Solution structure of AVR-Pia | | Descriptor: | AVR-Pia protein | | Authors: | Ose, T, Oikawa, A, Nakamura, Y, Maenaka, K, Higuchi, Y, Satoh, Y, Fujiwara, S, Demura, M, Sone, T. | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae
J.Biomol.Nmr, 63, 2015
|
|
2N39
 
 | |
2N3A
 
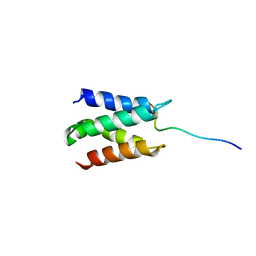 | | Solution structure of LEDGF/p75 IBD in complex with POGZ peptide (1389-1404) | | Descriptor: | PC4 and SFRS1-interacting protein, Pogo transposable element with ZNF domain | | Authors: | Tesina, P, Cermakova, K, Horejsi, M, Prochazkova, K, Fabry, M, Sharma, S, Christ, F, Demeulemeester, J, Debyser, Z, De Rijck, J, Veverka, V, Rezacova, P. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif.
Nat Commun, 6, 2015
|
|
2N3B
 
 | | Structure of oxidized horse heart cytochrome c encapsulated in reverse micelles | | Descriptor: | Cytochrome c, HEME C | | Authors: | O'Brien, E.S, Nucci, N.V, Fuglestad, B, Tommos, C, Wand, A. | | Deposit date: | 2015-05-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Defining the Apoptotic Trigger: THE INTERACTION OF CYTOCHROME c AND CARDIOLIPIN.
J.Biol.Chem., 290, 2015
|
|
2N3D
 
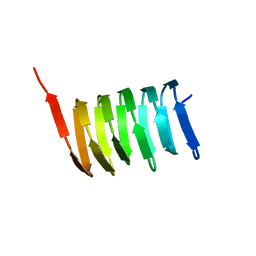 | | Atomic structure of the cytoskeletal bactofilin BacA revealed by solid-state NMR | | Descriptor: | Bactofilin A | | Authors: | Shi, C, Fricke, P, Lin, L, Chevelkov, V, Wegstroth, M, Giller, K, Becker, S, Thanbichler, M, Lange, A. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR.
Sci Adv, 1, 2015
|
|
2N3E
 
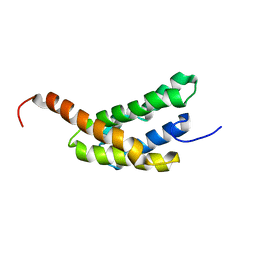 | | Amino-terminal domain of Latrodectus hesperus MaSp1 with neutralized acidic cluster | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Schaal, D, Bauer, J, Schweimer, K, Scheibel, T, Roesch, P, Schwarzinger, S. | | Deposit date: | 2015-05-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of an engineered amino-terminal ampullate spider silk with neutralized charge cluster
To be Published
|
|
2N3F
 
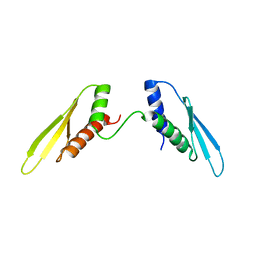 | |
2N3G
 
 | |
2N3H
 
 | |
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | Descriptor: | Heat shock protein beta-1 | | Authors: | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
2N3K
 
 | | Human Brd4 ET domain in complex with MLV Integrase C-term | | Descriptor: | Bromodomain-containing protein 4, MLV integrase | | Authors: | Crowe, B.L, Foster, M.P. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Brd4 ET domain bound to a C-terminal motif from gamma-retroviral integrases reveals a conserved mechanism of interaction.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N3L
 
 | |
2N3M
 
 | |
2N3O
 
 | | Structure of PTB RRM1(41-163) bound to an RNA stemloop containing a structured loop derived from viral internal ribosomal entry site RNA | | Descriptor: | Polypyrimidine tract-binding protein 1, RNA (5'-R(*GP*GP*GP*AP*CP*CP*UP*GP*GP*UP*CP*UP*UP*UP*CP*CP*AP*GP*GP*UP*CP*CP*C)-3') | | Authors: | Maris, C, Jayne, S.F, Damberger, F.F, Ravindranathan, S, Allain, F.H.-T. | | Deposit date: | 2015-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | C-terminal helix folding upon pyrimidine-rich hairpin binding to PTB RRM1. Implications for PTB function in Encephalomyocarditis virus IRES activity.
To be Published
|
|
2N3P
 
 | |
2N3Q
 
 | |
2N3R
 
 | | NMR structure of the II-III-VI three-way junction from the VS ribozyme and identification of magnesium-binding sites using paramagnetic relaxation enhancement | | Descriptor: | MAGNESIUM ION, RNA (62-MER) | | Authors: | Bonneau, E, Girard, N, Lemieux, S, Legault, P. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the II-III-VI three-way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure.
Rna, 21, 2015
|
|
2N3S
 
 | |
2N3T
 
 | |
2N3U
 
 | |
2N3V
 
 | |
