2MIA
 
 | |
2MIC
 
 | |
2MID
 
 | |
2MIE
 
 | |
2MIF
 
 | |
2MIG
 
 | |
2MIH
 
 | |
2MII
 
 | | NMR structure of E. coli LpoB | | Descriptor: | Penicillin-binding protein activator LpoB | | Authors: | Jean, N.L, Egan, A.J.F, Koumoutsi, A, Bougault, C.M, Typas, A, Vollmer, W, Simorre, J.P. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MIJ
 
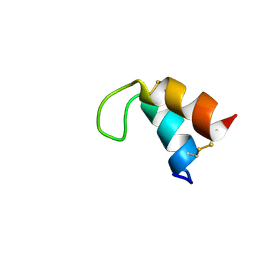 | | NMR structure of the S-linked glycopeptide sublancin 168 | | Descriptor: | SPBc2 prophage-derived bacteriocin sublancin-168, beta-D-glucopyranose | | Authors: | Garcia De Gonzalo, C.V, Zhu, L, Oman, T.J, van der Donk, W.A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the s-linked glycopeptide sublancin 168.
Acs Chem.Biol., 9, 2014
|
|
2MIM
 
 | |
2MIO
 
 | | Solution NMR Structure of SH3 Domain 1 of Rho GTPase-activating Protein 10 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9129A | | Descriptor: | Rho GTPase-activating protein 10 | | Authors: | Xu, X, Eletsky, A, Pulavarti, S.V.S.R.K, Wang, H, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of SH3 Domain 1 of Rho GTPase-activating Protein 10 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9129A
To be Published
|
|
2MIQ
 
 | | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 of Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J | | Descriptor: | Lysine-specific demethylase lid, ZINC ION | | Authors: | Xu, X, Eletsky, A, Shastry, R, Maglaqui, M, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 from Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J
To be Published
|
|
2MIS
 
 | |
2MIT
 
 | |
2MIU
 
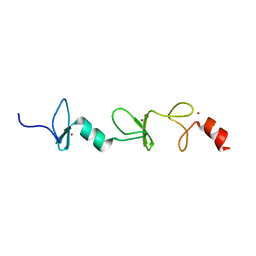 | | Structure of FHL2 LIM adaptor and its Interaction with Ski | | Descriptor: | Four and a half LIM domains protein 2, ZINC ION | | Authors: | Yang, Y, Sun, Y, Medrano, E.E, Tian, X, Weiss, M.A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of FHL2 LIM adaptor and its Interaction with Ski
To be Published
|
|
2MIV
 
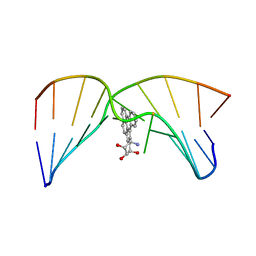 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2MIW
 
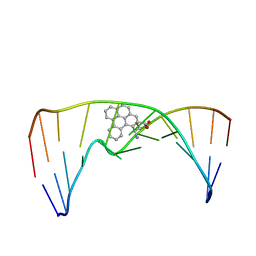 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2MIX
 
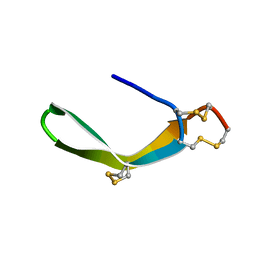 | | Structure of a novel venom peptide toxin from sample limited terebrid marine snail | | Descriptor: | venom peptide toxin | | Authors: | Bhuiyan, M.H, Anand, P, Grigoryan, A, Holford, M, Poget, S.F. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sample limited characterization of a novel disulfide-rich venom peptide toxin from terebrid marine snail Terebra variegata.
Plos One, 9, 2014
|
|
2MIY
 
 | |
2MIZ
 
 | |
2MJ0
 
 | |
2MJ1
 
 | |
2MJ2
 
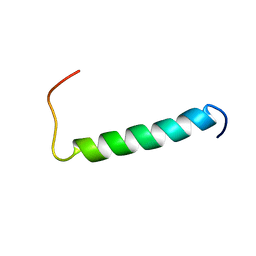 | | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, S.A, Abou-Gharbia, M, Childers, W, White, M, Bouaziz, S, Safak, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix
J.Virol., 2014
|
|
2MJ3
 
 | |
2MJ4
 
 | |
