2KIL
 
 | |
2KIM
 
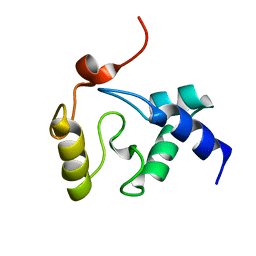 | | 1.7-mm microcryoprobe solution NMR structure of an O6-methylguanine DNA methyltransferase family protein from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium target VpR247. | | Descriptor: | O6-methylguanine-DNA methyltransferase | | Authors: | Aramini, J.M, Belote, R.L, Ciccosanti, C.T, Jiang, M, Rost, B, Nair, R, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of an O6-methylguanine DNA methyltransferase family protein from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium target VpR247.
To be Published
|
|
2KIO
 
 | |
2KIQ
 
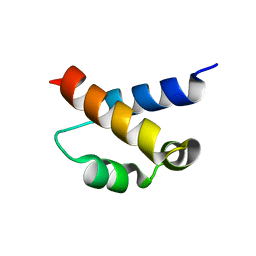 | | Solution structure of the FF Domain 2 of human transcription elongation factor CA150 | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Zeng, J, Boyles, J, Tripathy, C, Yan, A, Zhou, P, Donald, B.R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution protein structure determination starting with a global fold calculated from exact solutions to the RDC equations.
J.Biomol.Nmr, 45, 2009
|
|
2KIR
 
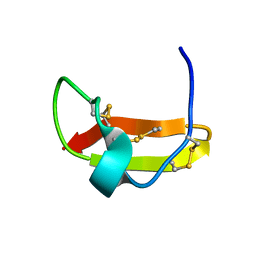 | | Solution structure of a designer toxin, mokatoxin-1 | | Descriptor: | Designer toxin | | Authors: | Biancalana, M, Koide, A, Takacs, Z, Goldstein, S, Koide, S. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-29 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | A designer ligand specific for Kv1.3 channels from a scorpion neurotoxin-based library.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KIS
 
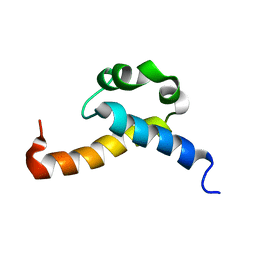 | | Solution structure of CA150 FF1 domain and FF1-FF2 interdomain linker | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Murphy, J.M, Hansen, D, Wiesner, S, Muhandiram, D, Borg, M, Smith, M.J, Sicheri, F, Kay, L.E, Forman-Kay, J.D, Pawson, T. | | Deposit date: | 2009-05-08 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays.
J.Mol.Biol., 393, 2009
|
|
2KIT
 
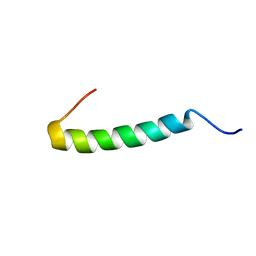 | |
2KIU
 
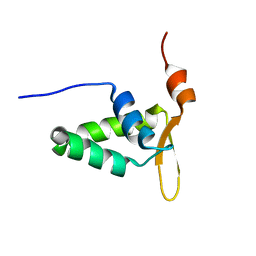 | |
2KIV
 
 | | AIDA-1 SAM domain tandem | | Descriptor: | Ankyrin repeat and sterile alpha motif domain-containing protein 1B | | Authors: | Donaldson, L.W, Kurabi, A. | | Deposit date: | 2009-05-12 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A nuclear localization signal at the SAM-SAM domain interface of AIDA-1 suggests a requirement for domain uncoupling prior to nuclear import.
J.Mol.Biol., 392, 2009
|
|
2KIW
 
 | | Solution NMR structure of the domain N-terminal to the integrase domain of SH1003 from Staphylococcus haemolyticus. Northeast Structural Genomics Consortium Target ShR105F (64-166). | | Descriptor: | Int protein | | Authors: | Yang, Y, Ramelot, T.A, Belote, R.L, Foote, E.L, Janjua, H, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-12 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the domain N-terminal to the integrase domain of SH1003 from Staphylococcus
haemolyticus. Northeast Structural Genomics Consortium Target ShR105F (64-166).
To be Published
|
|
2KIX
 
 | |
2KIZ
 
 | | Solution structure of Arkadia RING-H2 finger domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Kandias, N.G, Chasapis, C.T, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase.
Proteins, 80, 2012
|
|
2KJ1
 
 | |
2KJ3
 
 | | High-resolution structure of the HET-s(218-289) prion in its amyloid form obtained by solid-state NMR | | Descriptor: | Small s protein | | Authors: | Van Melckebeke, H, Wasmer, C, Lange, A, AB, E, Loquet, A, Meier, B.H. | | Deposit date: | 2009-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of HET-s(218-289) Amyloid Fibrils by Solid-State NMR Spectroscopy
J.Am.Chem.Soc., 132, 2010
|
|
2KJ4
 
 | | Solution structure of the complex of VEK-30 and plasminogen kringle 2 | | Descriptor: | VEK-30, plasminogen | | Authors: | Wang, M, Zajicek, J, Prorok, M, Castellin, F.J. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex of VEK-30 and plasminogen kringle 2.
J.Struct.Biol., 169, 2010
|
|
2KJ5
 
 | | Solution NMR structure of a domain from a putative phage integrase protein Nmul_A0064 from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR46C | | Descriptor: | Phage integrase | | Authors: | Mills, J.L, Eletsky, A, Ghosh, A, Wang, D, Lee, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a domain from a putative phage integrase protein Nmul_A0064 from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR46C.
To be Published
|
|
2KJ6
 
 | | NMR Solution Structure of a Tubulin folding cofactor B obtained from Arabidopsis thaliana: Northeast Structural Genomics Consortium target AR3436A | | Descriptor: | Tubulin folding cofactor B | | Authors: | Mani, R, Swapna, G.V.T, Shastry, R, Foote, E, Ciccosanti, C, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y.J, Acton, T, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Tbulin folding Cofactor B obtained from Arabidopsis thaliana: Northeast
To be Published
|
|
2KJ7
 
 | | Three-Dimensional NMR Structure of Rat Islet Amyloid Polypeptide in DPC micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Nanga, R, Brender, J.R, Xu, J, Hartman, K, Subramanian, V, Ramamoorthy, A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy.
J.Am.Chem.Soc., 131, 2009
|
|
2KJ8
 
 | | NMR structure of fragment 87-196 from the putative phage integrase IntS of E. coli: Northeast Structural Genomics Consortium target ER652A, PSI-2 | | Descriptor: | Putative prophage CPS-53 integrase | | Authors: | Cort, J.R, Ramelot, T.A, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Swapna, G, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-25 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of fragment 87-196 from the putative phage integrase IntS of E. coli
To be Published
|
|
2KJ9
 
 | | NMR structure of IntB phage-integrase-like protein fragment 90-199 from Erwinia carotova subsp. atroseptica: Northeast Structural Genomics Consortium target EwR217E | | Descriptor: | Integrase | | Authors: | Cort, J.R, Ramelot, T.A, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-25 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of IntB phage-integrase-like protein fragment 90-199 from Erwinia carotova subsp. atroseptica
To be Published
|
|
2KJA
 
 | | Chemical shift assignments, constraints, and coordinates for CN5 scorpion toxin | | Descriptor: | Beta-toxin Cn5 | | Authors: | Prochnicka-Chalufour, A, Corzo, G, Garcia, B.I, Possani, L.D, Delepierre, M. | | Deposit date: | 2009-05-26 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Cn5, a crustacean toxin found in the venom of the scorpions Centruroides noxius and Centruroides suffusus suffusus.
Biochim.Biophys.Acta, 1794, 2009
|
|
2KJB
 
 | |
2KJC
 
 | |
2KJD
 
 | |
2KJE
 
 | | NMR structure of CBP TAZ2 and adenoviral E1A complex | | Descriptor: | CREB-binding protein, Early E1A 32 kDa protein, ZINC ION | | Authors: | Ferreon, J.C, Martinez-Yamout, M, Dyson, H, Wright, P.E. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
