1ZWV
 
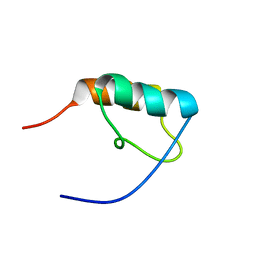 | |
1ZXA
 
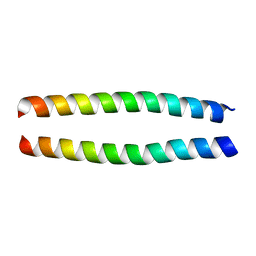 | | Solution Structure of the Coiled-Coil Domain of cGMP-dependent Protein Kinase Ia | | Descriptor: | cGMP-dependent protein kinase 1, alpha isozyme | | Authors: | Schnell, J.R, Zhou, G.P, Zweckstetter, M, Rigby, A.C, Chou, J.J. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rapid and accurate structure determination of coiled-coil domains using NMR dipolar couplings: Application to cGMP-dependent protein kinase I{alpha}
Protein Sci., 14, 2005
|
|
1ZXF
 
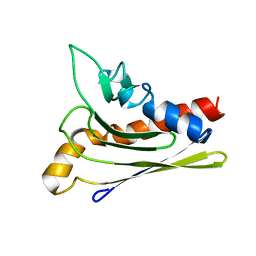 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
1ZXG
 
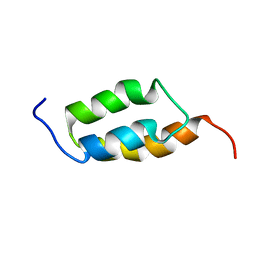 | | Solution structure of A219 | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | He, Y, Yeh, D.C, Alexander, P, Bryan, P.N, Orban, J. | | Deposit date: | 2005-06-08 | | Release date: | 2005-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of IgG binding domains with artificially evolved high levels of sequence identity but different folds.
Biochemistry, 44, 2005
|
|
1ZXH
 
 | | G311 mutant protein | | Descriptor: | Immunoglobulin G binding protein G | | Authors: | He, Y, Yeh, D.C, Alexander, P, Bryan, P.N, Orban, J. | | Deposit date: | 2005-06-08 | | Release date: | 2005-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of IgG binding domains with artificially evolved high levels of sequence identity but different folds.
Biochemistry, 44, 2005
|
|
1ZY3
 
 | |
1ZY6
 
 | | Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hairpin Antimicrobial Peptide in Lipid Bilayers from Rotational-Echo Double-Resonance Solid-State NMR | | Descriptor: | Protegrin 1 | | Authors: | Wu, X, Mani, R, Tang, M, Buffy, J.J, Waring, A.J, Sherman, M.A, Hong, M. | | Deposit date: | 2005-06-09 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-Bound Dimer Structure of a beta-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR.
Biochemistry, 45, 2006
|
|
1ZYF
 
 | | Structure of a Supercoiling Responsive DNA Site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*AP*TP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
1ZYG
 
 | | Structure of a Supercoiling Responsive DNA Site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*CP*GP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
1ZYH
 
 | | Structure of a Supercoiling Responsive DNA site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*AP*GP*GP*GP*TP*TP*G)-3', 5'-D(*CP*AP*AP*CP*CP*CP*TP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
1ZZA
 
 | | Solution NMR Structure of the Membrane Protein Stannin | | Descriptor: | Stannin | | Authors: | Buck-Koehntop, B.A, Mascioni, A, Buffy, J.J, Veglia, G. | | Deposit date: | 2005-06-13 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and membrane topology of stannin: a mediator of neuronal cell apoptosis induced by trimethyltin chloride.
J.Mol.Biol., 354, 2005
|
|
1ZZF
 
 | |
1ZZV
 
 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter FecA from Escherichia coli. | | Descriptor: | Iron(III) dicitrate transport protein fecA | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-14 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
214D
 
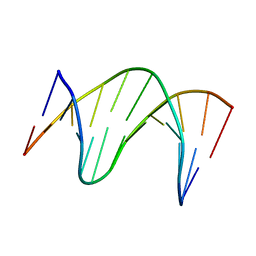 | | THE SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING A SINGLE 2'-O-METHYL-BETA-D-ARAT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*AP*TP*AP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*AP*(T41)P*AP*TP*GP*CP*G)-3') | | Authors: | Gotfredsen, C.H, Spielmann, H.P, Wengel, J, Jacobsen, J.P. | | Deposit date: | 1995-07-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA duplex containing a single 2'-O-methyl-beta-D-araT: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Bioconjug.Chem., 7, 1996
|
|
229D
 
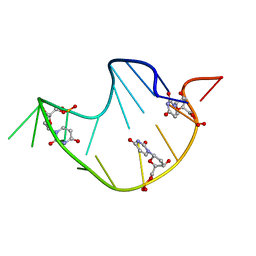 | | DNA ANALOG OF YEAST TRANSFER RNA PHE ANTICODON DOMAIN WITH MODIFIED BASES 5-METHYL CYTOSINE AND 1-METHYL GUANINE | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*CP*(UMP)P*GP*AP*AP*(MG1)P*AP*(UMP)P*(5CM)P*(UMP)P*GP*G)-3') | | Authors: | Basti, M.M, Stuart, J.W, Lam, A.T, Guenther, R, Agris, P.F. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design, biological activity and NMR-solution structure of a DNA analogue of yeast tRNA(Phe) anticodon domain.
Nat.Struct.Biol., 3, 1996
|
|
28SP
 
 | |
2A00
 
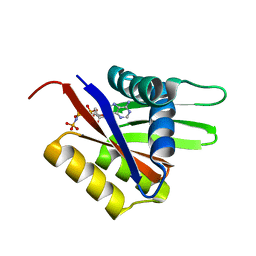 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
2A02
 
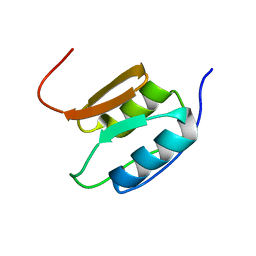 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter PupA from Pseudomonas putida. | | Descriptor: | Ferric-pseudobactin 358 receptor | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-15 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
2A05
 
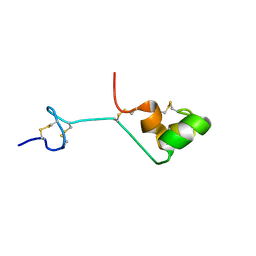 | | The cysteine-rich secretory protein domain of Tpx-1 is related to ion channel toxins and regulates Ryanodine receptor Ca2+ signaling | | Descriptor: | Cysteine-rich secretory protein-2 | | Authors: | Gibbs, G.M, Scanlon, M.J, Swarbrick, J, Curtis, S, Dulhunty, A.F, O'Bryan, M.K. | | Deposit date: | 2005-06-15 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The cysteine-rich secretory protein domain of Tpx-1 is related to ion channel toxins and regulates ryanodine receptor Ca2+ signaling.
J.Biol.Chem., 281, 2006
|
|
2A0A
 
 | |
2A0T
 
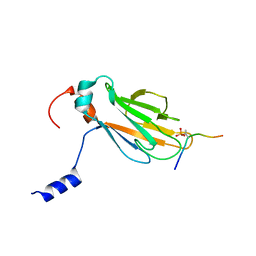 | | NMR structure of the FHA1 domain of Rad53 in complex with a biological relevant phosphopeptide derived from Madt1 | | Descriptor: | Hypothetical 73.8 kDa protein in SAS3-SEC17 intergenic region, residues 301-310, Serine/threonine-protein kinase RAD53 | | Authors: | Mahajan, A, Yuan, C, Pike, B.L, Heierhorst, J, Chang, C.-F, Tsai, M.-D. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | FHA Domain-Ligand Interactions: Importance of Integrating Chemical and Biological Approaches
J.Am.Chem.Soc., 127, 2005
|
|
2A1C
 
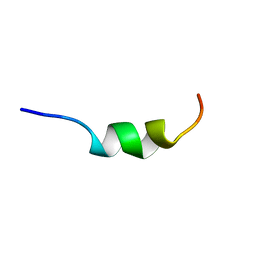 | | Solution structure of CSP1 | | Descriptor: | CSP1 | | Authors: | Johnsborg, O, Kristiansen, P.E. | | Deposit date: | 2005-06-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Hydrophobic Patch in the Pneumococcal Competence Pheromone CSP is Essential for Specificity and Biological Activity
To be Published
|
|
2A20
 
 | | Solution structure of Rim2 Zinc Finger Domain | | Descriptor: | Regulating synaptic membrane exocytosis protein 2, ZINC ION | | Authors: | Dulubova, I, Lou, X, Lu, J, Huryeva, I, Alam, A, Schneggenburger, R, Sudhof, T.C, Rizo, J. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity?
Embo J., 24, 2005
|
|
2A24
 
 | | HADDOCK Structure of HIF-2a/ARNT PAS-B Heterodimer | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain protein 1 | | Authors: | Card, P.B, Erbel, P.J, Gardner, K.H. | | Deposit date: | 2005-06-21 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization.
J.Mol.Biol., 353, 2005
|
|
2A2B
 
 | | Curvacin A | | Descriptor: | Bacteriocin curvacin A | | Authors: | Haugen, H.S, Kristiansen, P.E. | | Deposit date: | 2005-06-22 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide curvacin A
Biochemistry, 44, 2005
|
|
