1X5V
 
 | | NMR Structure of PcFK1 | | Descriptor: | PcFK1 | | Authors: | Pimentel, C, Choi, S.J, Chagot, B, Guette, C, Camadro, J.M, Darbon, H. | | Deposit date: | 2005-05-17 | | Release date: | 2006-04-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PcFK1, a spider peptide active against Plasmodium falciparum
Protein Sci., 15, 2006
|
|
1X60
 
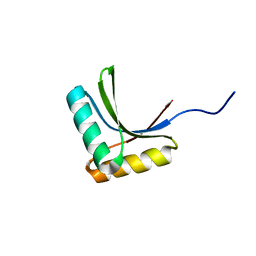 | | Solution structure of the peptidoglycan binding domain of B. subtilis cell wall lytic enzyme CwlC | | Descriptor: | Sporulation-specific N-acetylmuramoyl-L-alanine amidase | | Authors: | Mishima, M, Shida, T, Yabuki, K, Kato, K, Sekiguchi, J, Kojima, C. | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Peptidoglycan Binding Domain of Bacillus subtilis Cell Wall Lytic Enzyme CwlC: Characterization of the Sporulation-Related Repeats by NMR(,)
Biochemistry, 44, 2005
|
|
1X61
 
 | |
1X62
 
 | |
1X63
 
 | |
1X64
 
 | |
1X65
 
 | | Solution structure of the third cold-shock domain of the human KIAA0885 protein (UNR PROTEIN) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
1X66
 
 | | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor | | Descriptor: | Friend leukemia integration 1 transcription factor | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor
To be Published
|
|
1X67
 
 | | Solution structure of the cofilin homology domain of HIP-55 (drebrin-like protein) | | Descriptor: | Drebrin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
1X69
 
 | | Solution structures of the SH3 domain of human Src substrate cortactin | | Descriptor: | cortactin isoform a | | Authors: | Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domain of human Src substrate cortactin
To be Published
|
|
1X6B
 
 | | Solution structures of the SH3 domain of human rho guanine exchange factor (GEF) 16 | | Descriptor: | Rho guanine exchange factor (GEF) 16 | | Authors: | Sato, M, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domain of human rho guanine exchange factor (GEF) 16
To be Published
|
|
1X6C
 
 | | Solution structures of the SH2 domain of human protein-tyrosine phosphatase SHP-1 | | Descriptor: | Tyrosine-protein phosphatase, non-receptor type 6 | | Authors: | Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH2 domain of human protein-tyrosine phosphatase SHP-1
To be Published
|
|
1X6D
 
 | | Solution structures of the PDZ domain of human Interleukin-16 | | Descriptor: | Interleukin-16 | | Authors: | Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the PDZ domain of human Interleukin-16
To be Published
|
|
1X6E
 
 | | Solution structures of the C2H2 type zinc finger domain of human Zinc finger protein 24 | | Descriptor: | ZINC ION, Zinc finger protein 24 | | Authors: | Sato, M, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C2H2 type zinc finger domain of human Zinc finger protein 24
To be Published
|
|
1X6F
 
 | | Solution structures of the C2H2 type zinc finger domain of human Zinc finger protein 462 | | Descriptor: | ZINC ION, Zinc finger protein 462 | | Authors: | Sato, M, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C2H2 type zinc finger domain of human Zinc finger protein 462
To be Published
|
|
1X6G
 
 | | Solution structures of the SH3 domain of human megakaryocyte-associated tyrosine-protein kinase. | | Descriptor: | Megakaryocyte-associated tyrosine-protein kinase | | Authors: | Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domain of human megakaryocyte-associated tyrosine-protein kinase.
To be Published
|
|
1X6H
 
 | | Solution structures of the C2H2 type zinc finger domain of human Transcriptional repressor CTCF | | Descriptor: | Transcriptional repressor CTCF, ZINC ION | | Authors: | Sato, M, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C2H2 type zinc finger domain of human Transcriptional repressor CTCF
To be Published
|
|
1X7K
 
 | |
1X9A
 
 | | Solution NMR Structure of Protein Tm0979 from Thermotoga maritima. Ontario Center for Structural Proteomics Target TM0979_1_87; Northeast Structural Genomics Consortium Target VT98. | | Descriptor: | hypothetical protein TM0979 | | Authors: | Gaspar, J.A, Liu, C, Vassall, K.A, Stathopulos, P.B, Meglei, G, Stephen, R, Pineda-Lucena, A, Wu, B, Yee, A, Arrowsmith, C.H, Meiering, E.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel member of the YchN-like fold: solution structure of the hypothetical protein Tm0979 from Thermotoga maritima.
Protein Sci., 14, 2005
|
|
1X9B
 
 | | Solution NMR Structure of Protein Ta0354 from Thermoplasma acidophilum. Ontario Center for Structural Proteomics target TA0354_69_121; Northeast Structural Genomics Consortium Target TaT38. | | Descriptor: | hypothetical membrane protein ta0354_69_121 | | Authors: | Wu, B, Yee, A, Huang, Y.J, Ramelot, T.A, Semesi, A, Lemak, A, Edward, A, Kennedy, M, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical membrane protein ta0354_69_121 from Thermoplasma acidophilum
To be Published
|
|
1X9L
 
 | | Solution structure of CuI-DR1885 from Deinococcus Radiodurans | | Descriptor: | COPPER (I) ION, CuI-DR1885 | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Katsari, E, Katsaros, N, Kubicek, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-08-23 | | Release date: | 2004-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1X9V
 
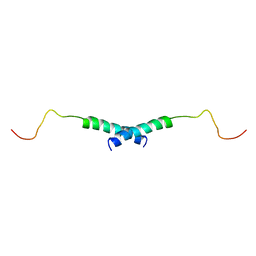 | | Dimeric structure of the C-terminal domain of Vpr | | Descriptor: | VPR protein | | Authors: | Bourbigot, S, Beltz, H, Denis, J, Morellet, N, Roques, B.P, Mely, Y, Bouaziz, S. | | Deposit date: | 2004-08-24 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of the HIV-1 regulatory protein Vpr adopts an antiparallel dimeric structure in solution via its leucine-zipper-like domain
Biochem.J., 387, 2005
|
|
1XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, 5 STRUCTURES | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-16 | | Release date: | 1999-07-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
1XBL
 
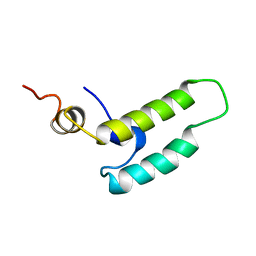 | | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | | Descriptor: | DNAJ | | Authors: | Pellecchia, M, Szyperski, T, Wall, D, Georgopoulos, C, Wuthrich, K. | | Deposit date: | 1996-10-07 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.
J.Mol.Biol., 260, 1996
|
|
1XC0
 
 | | Twenty Lowest Energy Structures of Pa4 by Solution NMR | | Descriptor: | Pardaxin P-4 | | Authors: | Porcelli, F, Buck, B, Lee, D.-K, Hallock, K.J, Ramamoorthy, A, Veglia, G. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of pardaxin determined by NMR experiments in model membranes
J.Biol.Chem., 279, 2004
|
|
