1NZM
 
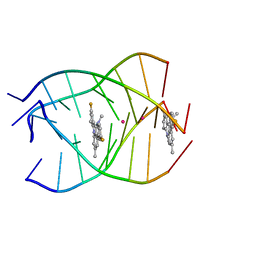 | | NMR structure of the parallel-stranded DNA quadruplex d(TTAGGGT)4 complexed with the telomerase inhibitor RHPS4 | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, 5'-D(*TP*TP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Gavathiotis, E, Heald, R.A, Stevens, M.F.G, Searle, M.S. | | Deposit date: | 2003-02-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Drug Recognition and Stabilisation of the Parallel-stranded DNA Quadruplex d(TTAGGGT)4 Containing the Human Telomeric Repeat
J.Mol.Biol., 334, 2003
|
|
1NZP
 
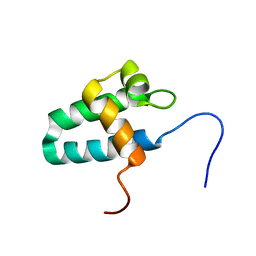 | | Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda | | Descriptor: | DNA polymerase lambda | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Bebenek, K, Garcia-Diaz, M, Blanco, L, Kunkel, T.A, London, R.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda
Biochemistry, 42, 2003
|
|
1O1W
 
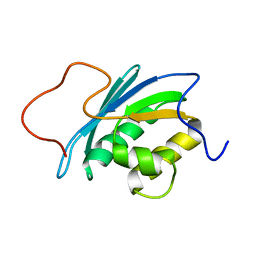 | | SOLUTION STRUCTURE OF THE RNASE H DOMAIN OF THE HIV-1 REVERSE TRANSCRIPTASE IN THE PRESENCE OF MAGNESIUM | | Descriptor: | RIBONUCLEASE H | | Authors: | Pari, K, Mueller, G.A, Derose, E.F, Kirby, T.W, London, R.E. | | Deposit date: | 2003-02-12 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Rnase H Domain of the HIV-1 Reverse Transcriptase in the Presence of Magnesium
Biochemistry, 42, 2003
|
|
1O4X
 
 | |
1O5P
 
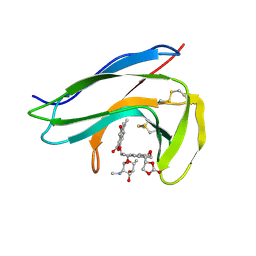 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
1O6X
 
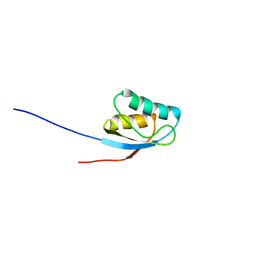 | | NMR solution structure of the activation domain of human procarboxypeptidase A2 | | Descriptor: | PROCARBOXYPEPTIDASE A2 | | Authors: | Jimenez, M.A, Villegas, V, Santoro, J, Serrano, L, Vendrell, J, Aviles, F.X, Rico, M. | | Deposit date: | 2002-10-17 | | Release date: | 2003-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Activation Domain of Human Procarboxypeptidase A2
Protein Sci., 12, 2003
|
|
1O78
 
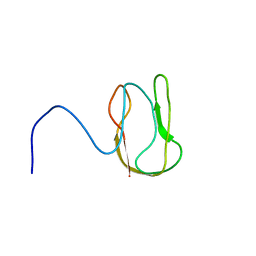 | |
1O7B
 
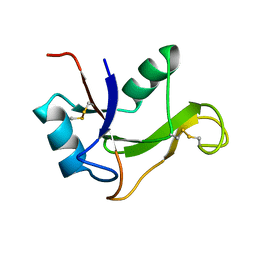 | | Refined solution structure of the human TSG-6 Link module | | Descriptor: | TUMOR NECROSIS FACTOR-INDUCIBLE PROTEIN TSG-6 | | Authors: | Blundell, C.D, Teriete, P, Kahmann, J.D, Pickford, A.R, Campbell, I.D, Day, A.J. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The link module from ovulation- and inflammation-associated protein TSG-6 changes conformation on hyaluronan binding.
J. Biol. Chem., 278, 2003
|
|
1O7C
 
 | | Solution structure of the human TSG-6 Link module in the presence of a hyaluronan octasaccharide | | Descriptor: | TUMOR NECROSIS FACTOR-INDUCIBLE PROTEIN TSG-6 | | Authors: | Blundell, C.D, Teriete, P, Kahmann, J.D, Pickford, A.R, Campbell, I.D, Day, A.J. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The link module from ovulation- and inflammation-associated protein TSG-6 changes conformation on hyaluronan binding.
J. Biol. Chem., 278, 2003
|
|
1O8R
 
 | |
1O8T
 
 | | Global Structure and Dynamics of Human Apolipoprotein CII in Complex with Micelles: Evidence for increased mobility of the helix involved in the activation of lipoprotein lipase | | Descriptor: | APOLIPOPROTEIN C-II | | Authors: | Zdunek, J, Martinez, G.V, Schleucher, J, Lycksell, P.O, Yin, Y, Nilsson, S, Shen, Y, Olivecrona, G, Wijmenga, S. | | Deposit date: | 2002-11-29 | | Release date: | 2003-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global Structure and Dynamics of Human Apolipoprotein Cii in Complex with Micelles: Evidence for Increased Mobility of the Helix Involved in the Activation of Lipoprotein Lipase
Biochemistry, 42, 2003
|
|
1O8Y
 
 | | Solution structure of SFTI-1(6,5), an acyclic permutant of the proteinase inhibitor SFTI-1, trans-trans-trans conformer (tt-A) | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Marx, U.C, Craik, D.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-03-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Enzymatic Cyclization of a Potent Bowman-Birk Protease Inhibitor, Sunflower Trypsin Inhibitor-1, and Solution Structure of an Acyclic Precursor Peptide
J.Biol.Chem., 278, 2003
|
|
1O8Z
 
 | | Solution structure of SFTI-1(6,5), an acyclic permutant of the proteinase inhibitor SFTI-1, cis-trans-trans conformer (ct-A) | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Marx, U.C, Craik, D.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-03-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Enzymatic Cyclization of a Potent Bowman-Birk Protease Inhibitor, Sunflower Trypsin Inhibitor-1, and Solution Structure of an Acyclic Precursor Peptide
J.Biol.Chem., 278, 2003
|
|
1OA5
 
 | |
1OA6
 
 | |
1OCA
 
 | | HUMAN CYCLOPHILIN A, UNLIGATED, NMR, 20 STRUCTURES | | Descriptor: | CYCLOPHILIN A | | Authors: | Ottiger, M, Zerbe, O, Guntert, P, Wuthrich, K. | | Deposit date: | 1997-07-07 | | Release date: | 1997-11-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution conformation of unligated human cyclophilin A.
J.Mol.Biol., 272, 1997
|
|
1OCD
 
 | | CYTOCHROME C (OXIDIZED) FROM EQUUS CABALLUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Qi, P.X, Beckman, R.A, Wand, A.J. | | Deposit date: | 1996-07-03 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of horse heart ferricytochrome c and detection of redox-related structural changes by high-resolution 1H NMR.
Biochemistry, 35, 1996
|
|
1OCI
 
 | | [3.2.0]bcANA:DNA | | Descriptor: | 5'-D(*CP*TP*GP*A TLBP*AP*TP*GP*CP)-3', 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*GP)-3' | | Authors: | Tommerholt, H.V, Christensen, N.K, Nielsen, P, Wengel, J, Stein, P.C, Jacobsen, J.P, Petersen, M. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a DsDNA Containing a Bicyclic D-Arabino-Configured Nucleotide Fixed in an O4'-Endo Sugar Conformation
Org.Biomol.Chem., 1, 2003
|
|
1OCP
 
 | | SOLUTION STRUCTURE OF OCT3 POU-HOMEODOMAIN | | Descriptor: | OCT-3 | | Authors: | Morita, E.H, Hayashi, F, Shirakawa, M, Kyogoku, Y. | | Deposit date: | 1995-02-21 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Oct-3 POU-Homeodomain in Solution, as Determined by Triple Resonance Heteronuclear Multidimensional NMR Spectroscopy
Protein Sci., 4, 1995
|
|
1ODP
 
 | |
1ODQ
 
 | |
1ODR
 
 | |
1OEF
 
 | | PEPTIDE OF HUMAN APOE RESIDUES 263-286, NMR, 5 STRUCTURES AT PH 4.8, 37 DEGREES CELSIUS AND PEPTIDE:SDS MOLE RATIO OF 1:90 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Wang, G, Pierens, G.K, Treleaven, W.D, Sparrow, J.T, Cushley, R.J. | | Deposit date: | 1996-03-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformations of human apolipoprotein E(263-286) and E(267-289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR.
Biochemistry, 35, 1996
|
|
1OEG
 
 | | PEPTIDE OF HUMAN APOE RESIDUES 267-289, NMR, 5 STRUCTURES AT PH 6.0, 37 DEGREES CELSIUS AND PEPTIDE:SDS MOLE RATIO OF 1:90 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Wang, G, Pierens, G.K, Treleaven, W.D, Sparrow, J.T, Cushley, R.J. | | Deposit date: | 1996-03-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformations of human apolipoprotein E(263-286) and E(267-289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR.
Biochemistry, 35, 1996
|
|
1OG7
 
 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-04-25 | | Release date: | 2003-09-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
