1N0C
 
 | | Stability of cyclic beta-hairpins: Asymmetric contibutions from side chains of hydrogen bonded cross-strand residue pair | | Descriptor: | bhp_HWLV, disulfide cyclized beta-hairpin peptide | | Authors: | Russel, S.J, Blandl, T, Skelton, N.J, Cochran, A.G. | | Deposit date: | 2002-10-11 | | Release date: | 2003-10-21 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Stability of cyclic beta-hairpins: asymmetric contributions from side chains of
a hydrogen-bonded cross-strand residue pair
J.AM.CHEM.SOC., 125, 2003
|
|
1N0D
 
 | | Stability of cyclic beta-hairpins: Asymmetric contibutions from side chains of hydrogen bonded cross-strand residue pair | | Descriptor: | bhp_VWLH, disulfide cyclized beta-hairpin peptide | | Authors: | Russell, S.J, Blandl, T, Skelton, N.J, Cochran, A.G. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Stability of Cyclic Beta-Hairpins: Asymmetric Contributions From Side Chains of Hydrogen Bonded Cross-Strand Residue Pair
J.Am.Chem.Soc., 125, 2003
|
|
1N0Z
 
 | | Solution structure of the first zinc-finger domain from ZNF265 | | Descriptor: | ZINC ION, ZNF265 | | Authors: | Plambeck, C.A, Fairley, K, Kwan, A.H.Y, Westman, B.J, Adams, D, Morris, B, Mackay, J.P. | | Deposit date: | 2002-10-15 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the zinc finger domain from human splicing factor ZNF265 fold
J.BIOL.CHEM., 278, 2003
|
|
1N1U
 
 | | NMR structure of [Ala1,15]kalata B1 | | Descriptor: | kalata B1 | | Authors: | Daly, N.L, Clark, R.J, Craik, D.J. | | Deposit date: | 2002-10-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Disulfide Folding Pathways of Cystine Knot Proteins. TYING THE KNOT WITHIN THE CIRCULAR BACKBONE OF THE CYCLOTIDES
J.Biol.Chem., 278, 2003
|
|
1N2Y
 
 | | SOLUTION STRUCTURE OF SS-CYCLIZED CATESTATIN FRAGMENT FROM CHROMOGRANIN A | | Descriptor: | CATESTATIN | | Authors: | Preece, N.E, Nguyen, M, Mahata, M, Mahata, S.K, Mahapatra, N.R, Tsigelny, I, O'Connor, D.T. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences and activities of peptides from the catecholamine
release-inhibitory (catestatin) region of chromogranin A
Regul.Pept., 118, 2004
|
|
1N3G
 
 | |
1N4B
 
 | | Solution Structure of the undecamer CGAAAC*TTTCG | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*D00*TP*TP*TP*CP*G)-3' | | Authors: | Webba da Silva, M, Noronha, A.M, Noll, D.M, Miller, P.S, Colvin, O.M, Gamcsik, M.P. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing
Mispair-Aligned N4C-Ethyl-N4C
Interstrand Cross-Linked Cytosines
Biochemistry, 41, 2002
|
|
1N4C
 
 | | NMR Structure of the J-Domain and Clathrin Substrate Binding Domain of Bovine Auxilin | | Descriptor: | Auxilin | | Authors: | Gruschus, J.M, Han, C.J, Greener, T, Greene, L.E, Ferretti, J.A, Eisenberg, E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles.
Biochemistry, 43, 2004
|
|
1N4I
 
 | | Solution structure of spruce budworm antifreeze protein at 5 degrees celsius | | Descriptor: | thermal hysteresis protein | | Authors: | Graether, S.P, Gagne, S.M, Spyracopoulos, L, Jia, Z, Davies, P.L, Sykes, B.D. | | Deposit date: | 2002-10-31 | | Release date: | 2003-04-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Spruce Budworm Antifreeze Protein: Changes in Structure and Dynamics at Low Temperature
J.Mol.Biol., 327, 2003
|
|
1N4N
 
 | | Structure of the Plant Defensin PhD1 from Petunia Hybrida | | Descriptor: | floral defensin-like protein 1 | | Authors: | Janssen, B.J.C, Schirra, H.J, Lay, F.T, Anderson, M.A, Craik, D.J. | | Deposit date: | 2002-11-01 | | Release date: | 2003-11-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of Petunia hybrida defensin 1, a novel plant defensin with five disulfide bonds
Biochemistry, 42, 2003
|
|
1N4Y
 
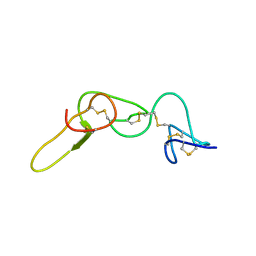 | | REFINED STRUCTURE OF KISTRIN | | Descriptor: | KISTRIN | | Authors: | Krezel, A.M, Krane, J, Dennis, M.S, Lazarus, R.A, Wagner, G. | | Deposit date: | 2002-11-02 | | Release date: | 2003-01-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist.
Science, 253, 1991
|
|
1N53
 
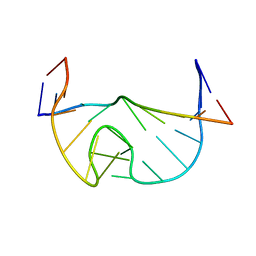 | | SOLUTION STRUCTURE OF B. SUBTILIS T BOX ANTITERMINATOR RNA | | Descriptor: | RNA (5'-R(*GP*AP*GP*GP*GP*UP*GP*GP*AP*AP*CP*CP*GP*CP*GP*C)-3'), RNA (5'-R(*GP*CP*GP*UP*CP*CP*CP*UP*C)-3') | | Authors: | Gerdeman, M.S, Henkin, T.M, Hines, J.V. | | Deposit date: | 2002-11-04 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Bacillus subtilis T-box Antiterminator RNA: Seven Nucleotide
Bulge Characterized by Stacking and Flexibility
J.Mol.Biol., 326, 2003
|
|
1N5G
 
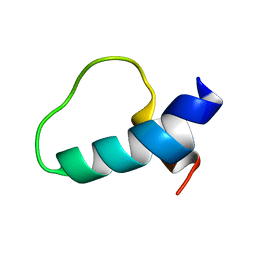 | |
1N66
 
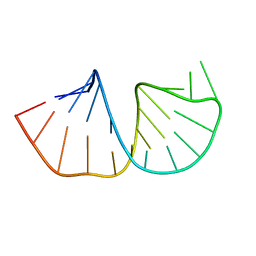 | | Structure of the pyrimidine-rich internal loop in the Y-domain of poliovirus 3'UTR | | Descriptor: | internal loop in the Y-domain of poliovirus 3'UTR | | Authors: | Lescrinier, E.M, Tessari, M, van Kuppeveld, F.J, Melchers, W.J, Hilbers, C.W, Heus, H.A. | | Deposit date: | 2002-11-08 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Pyrimidine-rich Internal Loop in the Poliovirus 3'-UTR: The Importance of Maintaining Pseudo-2-fold Symmetry in RNA Helices Containing Two Adjacent Non-canonical Base-pairs.
J.Mol.Biol., 331, 2003
|
|
1N6T
 
 | |
1N6U
 
 | | NMR structure of the interferon-binding ectodomain of the human interferon receptor | | Descriptor: | Interferon-alpha/beta receptor beta chain | | Authors: | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | Deposit date: | 2002-11-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
1N6V
 
 | | Average structure of the interferon-binding ectodomain of the human type I interferon receptor | | Descriptor: | Interferon-alpha/beta receptor beta chain | | Authors: | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | Deposit date: | 2002-11-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
1N6Z
 
 | |
1N7T
 
 | | ERBIN PDZ domain bound to a phage-derived peptide | | Descriptor: | 99-mer peptide of densin-180-like protein, phage-derived peptide | | Authors: | Skelton, N.J, Koehler, M.F.T, Zobel, K, Wong, W.L, Yeh, S, Pisabarro, M.T, Yin, J.P, Lasky, L.A, Sidhu, S.S. | | Deposit date: | 2002-11-16 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain.
J.Biol.Chem., 278, 2003
|
|
1N87
 
 | |
1N88
 
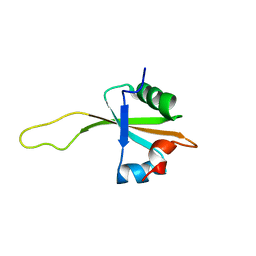 | | NMR structure of the ribosomal protein L23 from Thermus thermophilus. | | Descriptor: | Ribosomal protein L23 | | Authors: | Ohman, A, Rak, A, Dontsova, M, Garber, M.B, Hard, T. | | Deposit date: | 2002-11-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the ribosomal protein L23 from Thermus thermophilus.
J.Biomol.NMR, 26, 2003
|
|
1N89
 
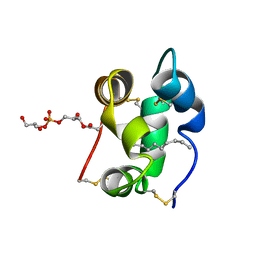 | | Solution structure of a liganded type 2 wheat non-specific Lipid Transfer Protein | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], lipid transfer protein | | Authors: | Pons, J.L, de Lamotte, F, Gautier, M.F, Delsuc, M.A. | | Deposit date: | 2002-11-20 | | Release date: | 2003-03-18 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of a liganded type 2 wheat nonspecific lipid transfer protein.
J.Biol.Chem., 278, 2003
|
|
1N8C
 
 | | Solution Structure of a Cis-Opened (10R)-N6-Deoxyadenosine Adduct of (9S,10R)-(9,10)-Epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA Duplex | | Descriptor: | (9S,10R)-9-HYDROXY-7,8,9,10-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*TP*CP*GP*TP*GP*AP*CP*CP*G)-3', 5'-D(*CP*GP*GP*TP*CP*AP*CP*GP*AP*GP*G)-3' | | Authors: | Volk, D.E, Thiviyanathan, V, Rice, J.S, Luxon, B.A, Shah, J.H, Yagi, H, Sayer, J.M, Yeh, H.J.C, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2002-11-20 | | Release date: | 2003-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Cis-Opened (10R)-N6-Deoxyadenosine Adduct of (9S,10R)-(9,10)-Epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA Duplex
Biochemistry, 42, 2003
|
|
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
1N8X
 
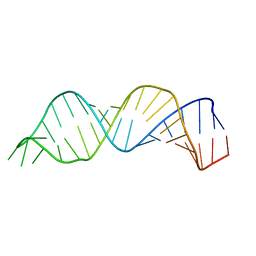 | | Solution structure of HIV-1 Stem Loop SL1 | | Descriptor: | HIV-1 STEM LOOP SL1 MONOMERIC RNA | | Authors: | Lawrence, D.C, Stover, C.C, Noznitsky, J, Wu, Z, Summers, M.F. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Intact Stem and Bulge of HIV-1 Psi-RNA Stem-Loop SL1
J.Mol.Biol., 326, 2003
|
|
