1LCC
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
1LCD
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
1LCM
 
 | |
1LCX
 
 | | NMR structure of HIV-1 gp41 659-671 13mer peptide | | Descriptor: | GP41 | | Authors: | Biron, Z, Khare, S, Samson, A.O, Hayek, Y, Naider, F, Anglister, J. | | Deposit date: | 2002-04-07 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Monomeric 3(10)-Helix Is Formed in Water by a 13-Residue Peptide
Representing the Neutralizing Determinant of HIV-1 on gp41.
Biochemistry, 41, 2002
|
|
1LD5
 
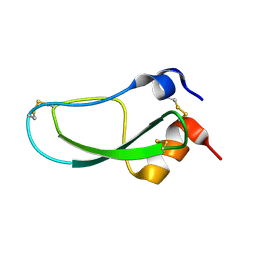 | | STRUCTURE OF BPTI MUTANT A16V | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR | | Authors: | Cierpicki, T, Otlewski, J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two variants of bovine pancreatic trypsin inhibitor (BPTI) reveal unexpected influence of mutations on protein structure and stability.
J.Mol.Biol., 321, 2002
|
|
1LD6
 
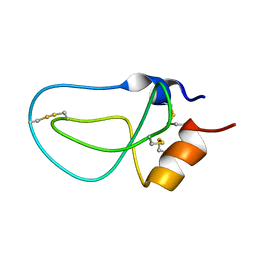 | | STRUCTURE OF BPTI_8A MUTANT | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR | | Authors: | Cierpicki, T, Otlewski, J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-09-11 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two variants of bovine pancreatic trypsin inhibitor (BPTI) reveal unexpected influence of mutations on protein structure and stability.
J.Mol.Biol., 321, 2002
|
|
1LDL
 
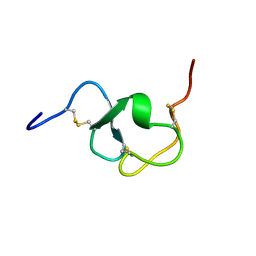 | |
1LDR
 
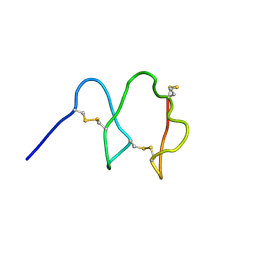 | |
1LDZ
 
 | |
1LEJ
 
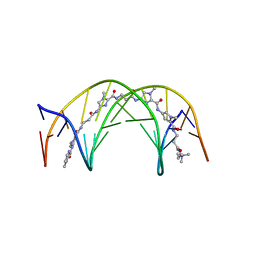 | | NMR Structure of a 1:1 Complex of Polyamide (Im-Py-Beta-Im-Beta-Im-Py-Beta-Dp) with the Tridecamer DNA Duplex 5'-CCAAAGAGAAGCG-3' | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*AP*GP*AP*AP*GP*CP*G)-3', 5'-D(*CP*GP*CP*TP*TP*CP*TP*CP*TP*TP*TP*GP*G)-3', IMIDAZOLE-PYRROLE-BETA ALANINE-IMIDAZOLE-BETA ALANINE-IMIDAZOLE-PYRROLE-BETA ALANINE-DIMETHYLAMINO PROPYLAMIDE | | Authors: | Urbach, A.R, Love, J.J, Ross, S.A, Dervan, P.B. | | Deposit date: | 2002-04-09 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a beta-alanine-linked polyamide bound to a full helical turn of purine tract DNA in the 1:1 motif.
J.Mol.Biol., 320, 2002
|
|
1LFC
 
 | | BOVINE LACTOFERRICIN (LFCINB), NMR, 20 STRUCTURES | | Descriptor: | LACTOFERRICIN | | Authors: | Hwang, P.M, Zhou, N, Shan, X, Arrowsmith, C.H, Vogel, H.J. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of lactoferricin B, an antimicrobial peptide derived from bovine lactoferrin.
Biochemistry, 37, 1998
|
|
1LFU
 
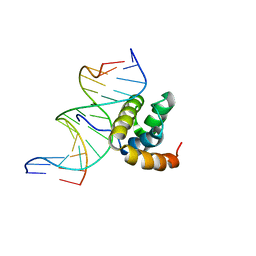 | | NMR Solution Structure of the Extended PBX Homeodomain Bound to DNA | | Descriptor: | 5'-D(*GP*CP*GP*CP*AP*TP*GP*AP*TP*TP*GP*CP*CP*C)-3', 5'-D(*GP*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*CP*GP*C)-3', homeobox protein PBX1 | | Authors: | Sprules, T, Green, N, Featherstone, M, Gehring, K. | | Deposit date: | 2002-04-12 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Lock and Key Binding of the HOX YPWM Peptide to the PBX Homeodomain
J.Biol.Chem., 278, 2003
|
|
1LG4
 
 | |
1LGL
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
1LIQ
 
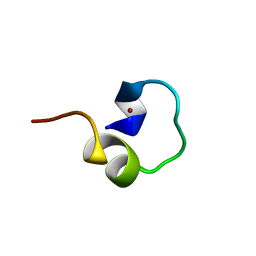 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
1LJV
 
 | | Bovine Pancreatic Polypeptide Bound to DPC Micelles | | Descriptor: | PANCREATIC HORMONE | | Authors: | Lerch, M, Gafner, V, Bader, R, Christen, B, Zerbe, O. | | Deposit date: | 2002-04-22 | | Release date: | 2002-10-09 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Bovine pancreatic polypeptide (bPP) undergoes significant changes in conformation and dynamics upon binding to DPC micelles.
J.Mol.Biol., 322, 2002
|
|
1LJZ
 
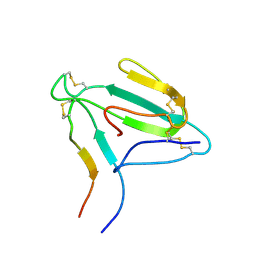 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Eisenstein, M, Chill, J.H, Anglister, J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1LKJ
 
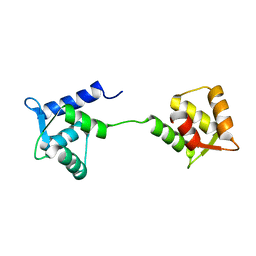 | | NMR Structure of Apo Calmodulin from Yeast Saccharomyces cerevisiae | | Descriptor: | Calmodulin | | Authors: | Ishida, H, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2002-04-25 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property.
Biochemistry, 41, 2002
|
|
1LKN
 
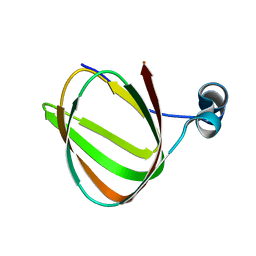 | |
1LKQ
 
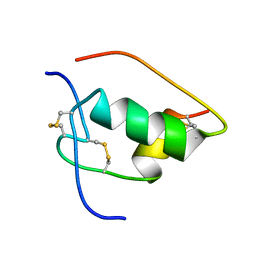 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-GLY, VAL-A3-GLY, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 20 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Weiss, M.A, Hua, Q.X, Chu, Y.C, Jia, W, Philips, N.F, Wang, R.Y, Katsoyannis, P.G. | | Deposit date: | 2002-04-25 | | Release date: | 2002-05-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Mechanism of insulin chain combination. Asymmetric roles of A-chain alpha-helices in disulfide pairing
J.Biol.Chem., 277, 2002
|
|
1LM0
 
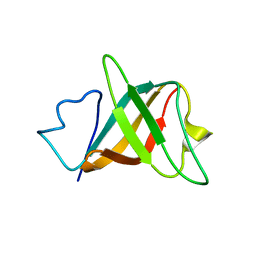 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
1LM2
 
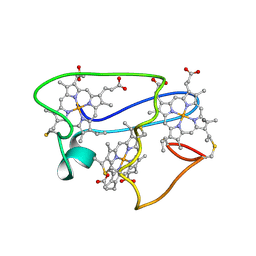 | | NMR structural characterization of the reduction of chromium(VI) to chromium(III) by cytochrome c7 | | Descriptor: | CHROMIUM ION, HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Bruschi, M, Michel, C, Turano, P. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The metal reductase activity of some multiheme cytochromes c: NMR structural characterization of the reduction of chromium(VI) to chromium(III) by cytochrome c(7).
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LMJ
 
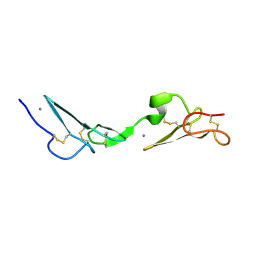 | | NMR Study of the Fibrillin-1 cbEGF12-13 Pair of Ca2+ Binding Epidermal Growth Factor-like Domains | | Descriptor: | CALCIUM ION, fibrillin 1 | | Authors: | Smallridge, R.S, Whiteman, P, Werner, J.M, Campbell, I.D, Handford, P.A, Downing, A.K. | | Deposit date: | 2002-05-02 | | Release date: | 2003-04-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of a Calcium Binding Epidermal Growth
Factor-like Domain Pair from the Neonatal Region of Human Fibrillin-1.
J.Biol.Chem., 278, 2003
|
|
1LMS
 
 | | Structural model for an alkaline form of ferricytochrome c | | Descriptor: | Cytochrome c, iso-1, HEME C | | Authors: | Assfalg, M, Bertini, I, Dolfi, A, Turano, P, Mauk, A.G, Rosell, F.I, Gray, H.B. | | Deposit date: | 2002-05-02 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural model for an alkaline form of ferricytochrome c
J.Am.Chem.Soc., 125, 2003
|
|
1LMV
 
 | |
