6BX9
 
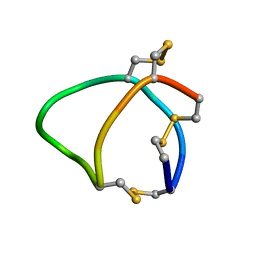 | | Solution structure of conotoxin reg3b | | Descriptor: | Conotoxin | | Authors: | Mari, F, Dovell, S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural plasticity of mini-M conotoxins - expression of all mini-M subtypes by Conus regius.
FEBS J., 285, 2018
|
|
6BY4
 
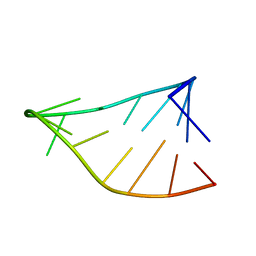 | | Single-State 14-mer UUCG Tetraloop calculated from Exact NOEs | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
6BY5
 
 | | Two-State 14-mer UUCG Tetraloop calculated from Exact NOEs (State one: Conformers 1-5, State Two: Conformers 6-10) | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
6BYV
 
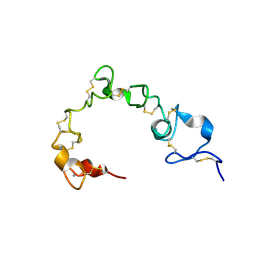 | | Solution NMR structure of cysteine-rich calcium bound domains of very low density lipoprotein receptor | | Descriptor: | CALCIUM ION, Very low-density lipoprotein receptor | | Authors: | Banerjee, K, Gruschus, J.M, Tjandra, N, Yakovlev, S, Medved, L. | | Deposit date: | 2017-12-21 | | Release date: | 2018-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Recombinant Fragment Containing Three Fibrin-Binding Cysteine-Rich Domains of the Very Low Density Lipoprotein Receptor.
Biochemistry, 57, 2018
|
|
6BZJ
 
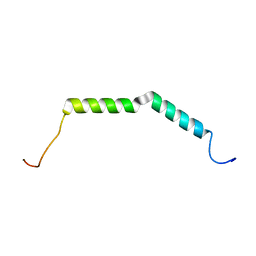 | | Solution structure of AGL55 | | Descriptor: | M protein | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
6BZK
 
 | | Solution structure of KTI55 | | Descriptor: | M protein | | Authors: | Castellino, F.J, Qiu, C, Yuan, Y. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
6BZL
 
 | | Solution structure of VEK75 | | Descriptor: | M protein | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
6C00
 
 | |
6C0A
 
 | | Actinin-1 EF-Hand bound to the Cav1.2 IQ Motif | | Descriptor: | Alpha-actinin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Turner, M.L, Ames, J.B. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Localization and Activation of Neuronal L-type Ca2+ Channels by a-Actinin1 and Calmodulin
To be Published
|
|
6C2U
 
 | | Solution structure of a phosphate-loop protein | | Descriptor: | phosphate-loop protein | | Authors: | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | Deposit date: | 2018-01-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C2V
 
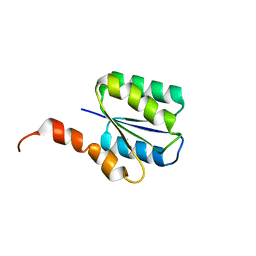 | | Solution structure of a phosphate-loop protein | | Descriptor: | phosphate-loop protein | | Authors: | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | Deposit date: | 2018-01-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C41
 
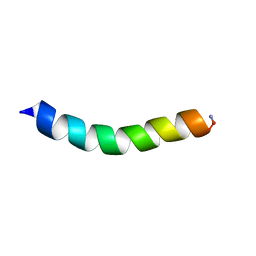 | |
6C44
 
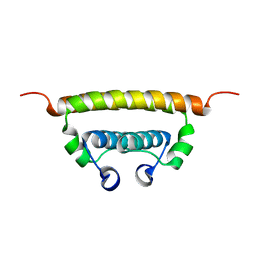 | | Zika virus capsid protein | | Descriptor: | Capsid protein | | Authors: | Morando, M.A, Barbosa, G.M, Cruz-Oliveira, C, Da Poian, A.T, Almeida, F.C.L. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamics of Zika Virus Capsid Protein in Solution: The Properties and Exposure of the Hydrophobic Cleft Are Controlled by the alpha-Helix 1 Sequence.
Biochemistry, 58, 2019
|
|
6C8U
 
 | | Solution structure of Musashi2 RRM1 | | Descriptor: | RNA-binding protein Musashi homolog 2 | | Authors: | Xing, M, Lan, L, Douglas, J.T, Gao, P, Hanzlik, R.P, Xu, L. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 2019
|
|
6CAH
 
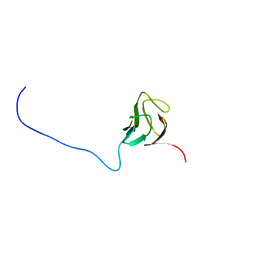 | |
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCJ
 
 | |
6CCW
 
 | |
6CCX
 
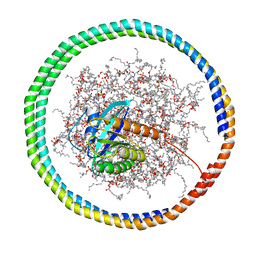 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CE5
 
 | |
6CEG
 
 | |
6CEI
 
 | |
6CEJ
 
 | |
6CFA
 
 | | peptide PaAMP1R3 | | Descriptor: | peptide PaAMP1R3 | | Authors: | Alves, E.S.F, Liao, L.M. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic peptide PaAMP1R3
To be Published
|
|
