6BF2
 
 | |
6BF3
 
 | | Solution structure of de novo macrocycle design7.3a | | Descriptor: | QDP(DPR)K(2TL)(DAS) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BF5
 
 | | Solution structure of de novo macrocycle design7.3a | | Descriptor: | QDP(DPR)K(DTH)(DAS) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BG9
 
 | |
6BGG
 
 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
6BGH
 
 | |
6BHN
 
 | | Red Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | Authors: | Yu, Q, Lim, S, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | Deposit date: | 2017-10-31 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BHO
 
 | | Green Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | Authors: | Lim, S, Yu, Q, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | Deposit date: | 2017-10-31 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BI5
 
 | |
6BI6
 
 | | Solution NMR structure of uncharacterized protein YejG | | Descriptor: | Uncharacterized protein YejG | | Authors: | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
6BJF
 
 | | NMR Structural and biophysical functional analysis of intracellular loop 5 of the NHE1 isoform of the Na+/H+ exchanger. | | Descriptor: | GLY-LEU-THR-TRP-PHE-ILE-ASN-LYS-PHE-ARG-ILE-VAL-LYS | | Authors: | McKay, R, Wong, K, Towle, K, Fliegel, L. | | Deposit date: | 2017-11-06 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Diverse residues of intracellular loop 5 of the Na+/H+exchanger modulate proton sensing, expression, activity and targeting.
Biochim Biophys Acta Biomembr, 1861, 2019
|
|
6BL9
 
 | |
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
6BP9
 
 | |
6BQI
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 2020
|
|
6BQS
 
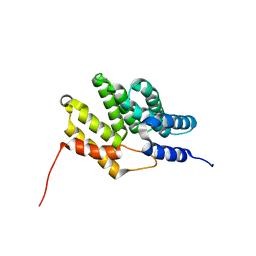 | | HusA haemophore from Porphyromonas gingivalis | | Descriptor: | hypothetical protein PG_2227 | | Authors: | Gell, D.A, Kwan, A.H, Horne, J, Hugrass, B.M, Collins, D.A.T. | | Deposit date: | 2017-11-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural properties of a haemophore facilitate targeted elimination of the pathogen Porphyromonas gingivalis.
Nat Commun, 9, 2018
|
|
6BR0
 
 | | Solution NMR structure for CcoTx-I | | Descriptor: | Beta-theraphotoxin-Cm1a | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2017-11-29 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Gating modifier toxins isolated from spider venom: Modulation of voltage-gated sodium channels and the role of lipid membranes.
J. Biol. Chem., 293, 2018
|
|
6BTV
 
 | | Solution NMR structures for CcoTx-II | | Descriptor: | Beta-theraphotoxin-Cm1b | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2017-12-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Gating modifier toxins isolated from spider venom: Modulation of voltage-gated sodium channels and the role of lipid membranes.
J. Biol. Chem., 293, 2018
|
|
6BUC
 
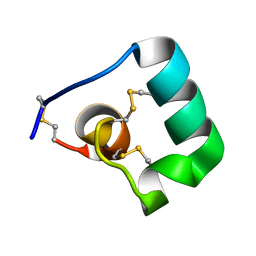 | |
6BUT
 
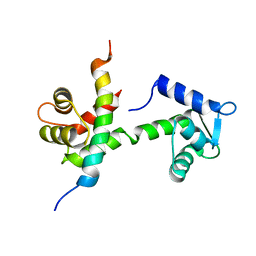 | |
6BV7
 
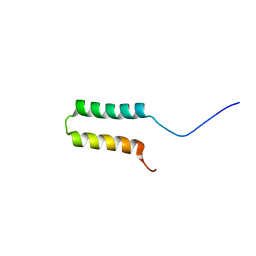 | |
6BVU
 
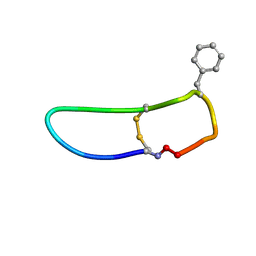 | | SFTI-HFRW-1 | | Descriptor: | Trypsin inhibitor 1 HFRW-1 | | Authors: | Schroeder, C.I. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6BVW
 
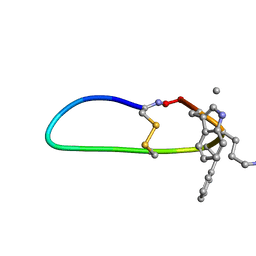 | | SFTI-HFRW-3 | | Descriptor: | Trypsin inhibitor 1 HFRW-3 | | Authors: | Schroeder, C.I. | | Deposit date: | 2017-12-14 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6BVX
 
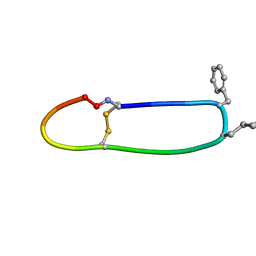 | | SFTI-HFRW-2 | | Descriptor: | Trypsin inhibitor 1 HFRW-2 | | Authors: | Schroeder, C.I. | | Deposit date: | 2017-12-14 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6BVY
 
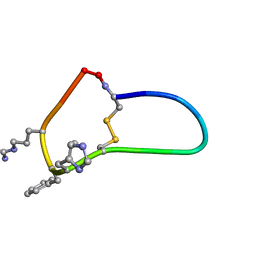 | | SFTI-HFRW-4 | | Descriptor: | Trypsin inhibitor 1 HFRW-4 | | Authors: | Schroeder, C.I, White, A. | | Deposit date: | 2017-12-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J. Med. Chem., 61, 2018
|
|
