5VKG
 
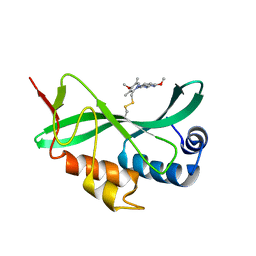 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with tenatoprazole | | Descriptor: | 4-methoxy-1-(5-methoxy-3H-imidazo[4,5-b]pyridin-2-yl)-3,5-dimethyl-2-(sulfanylmethyl)pyridin-1-ium, Tumor susceptibility gene 101 protein | | Authors: | Strickland, M, Ehrlich, L.S, Watanabe, S, Khan, M, Strub, M.-P, Luan, C.H, Powell, M.D, Leis, J, Tjandra, N, Carter, C. | | Deposit date: | 2017-04-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Tsg101 chaperone function revealed by HIV-1 assembly inhibitors.
Nat Commun, 8, 2017
|
|
5VKV
 
 | |
5VL6
 
 | |
5VLN
 
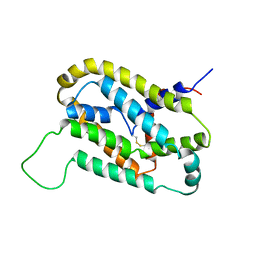
 | | NMR structure of the N-domain of troponin C bound to switch region of troponin I | | Descriptor: | Troponin C, slow skeletal and cardiac muscles,Troponin I, cardiac muscle | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures reveal details of small molecule binding to cardiac troponin.
J. Mol. Cell. Cardiol., 101, 2016
|
|
5VNT
 
 | |
5VO7
 
 | |
5VR1
 
 | |
5VR5
 
 | |
5VSO
 
 | | NMR structure of Ydj1 J-domain, a cytosolic Hsp40 from Saccharomyces cerevisiae | | Descriptor: | Yeast dnaJ protein 1 | | Authors: | Ciesielski, S.J, Tonelli, M, Lee, W, Cornilescu, G, Markley, J.L, Schilke, B.A, Ziegelhoffer, T, Craig, E.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Broadening the functionality of a J-protein/Hsp70 molecular chaperone system.
PLoS Genet., 13, 2017
|
|
5VTO
 
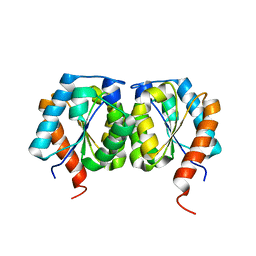 | |
5VWE
 
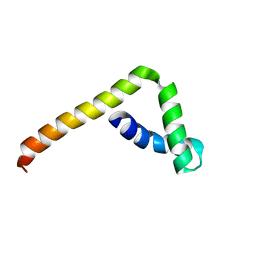 | |
5VWL
 
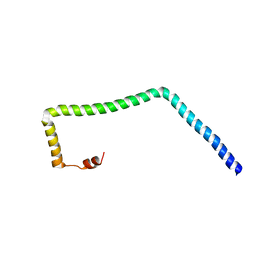 | |
5VX7
 
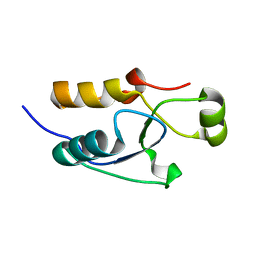 | |
5VZM
 
 | |
5W0Y
 
 | |
5W3G
 
 | |
5W3N
 
 | | Molecular structure of FUS low sequence complexity domain protein fibrils | | Descriptor: | RNA-binding protein FUS | | Authors: | Murray, D.T, Kato, M, Lin, Y, Thurber, K, Hung, I, McKnight, S, Tycko, R. | | Deposit date: | 2017-06-08 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains.
Cell, 171, 2017
|
|
5W4S
 
 | |
5W54
 
 | |
5W72
 
 | |
5W77
 
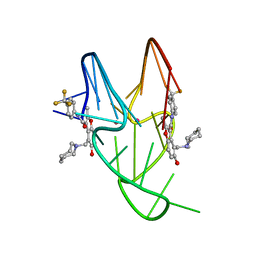 | | Solution structure of the MYC G-quadruplex bound to small molecule DC-34 | | Descriptor: | 4-[(azepan-1-yl)methyl]-5-hydroxy-2-methyl-N-[4-(trifluoromethyl)phenyl]-1-benzofuran-3-carboxamide, DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), POTASSIUM ION | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex.
Nat Commun, 9, 2018
|
|
5W88
 
 | |
5W8Y
 
 | |
5W8Z
 
 | |
5W96
 
 | | Solution structure of phage derived peptide inhibitor of frizzled 7 receptor | | Descriptor: | Fz7 binding peptide | | Authors: | Nile, A.H, de Sousa e Melo, F, Mukund, S, Piskol, R, Hansen, S, Zhou, L, Zhang, Y, Fu, Y, Gogol, E.B, Komuves, L.G, Modrusan, Z, Angers, S, Franke, Y, Koth, C, Fairbrother, W.J, Wang, W, de Sauvage, F.J, Hannoush, R.N. | | Deposit date: | 2017-06-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells.
Nat. Chem. Biol., 14, 2018
|
|
