1IKM
 
 | |
1IKU
 
 | | myristoylated recoverin in the calcium-free state, NMR, 22 structures | | Descriptor: | MYRISTIC ACID, RECOVERIN | | Authors: | Tanaka, T, Ames, J.B, Harvey, T.S, Stryer, L, Ikura, M. | | Deposit date: | 1996-01-18 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state.
Nature, 376, 1995
|
|
1IL6
 
 | | HUMAN INTERLEUKIN-6, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
1ILP
 
 | |
1IM1
 
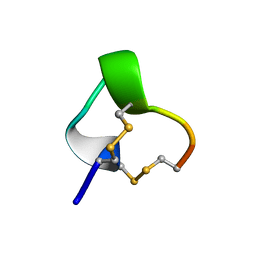 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
1IM7
 
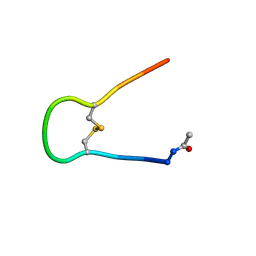 | | Solution structure of synthetic cyclic peptide mimicking the loop of HIV-1 gp41 glycoprotein envelope | | Descriptor: | GP41-PARENT PEPTIDE ACE-ILE-TRP-GLY-CYS-SER-GLY-LYS-LEU-ILE-CYS-THR-THR-ALA | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-05-10 | | Release date: | 2002-10-23 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1IML
 
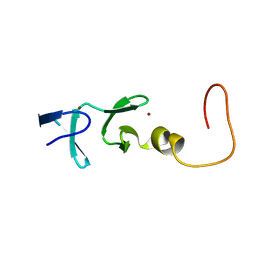 | | CYSTEINE RICH INTESTINAL PROTEIN, NMR, 48 STRUCTURES | | Descriptor: | CYSTEINE RICH INTESTINAL PROTEIN, ZINC ION | | Authors: | Perez-Alvarado, G.C, Kosa, J.L, Louis, H.A, Beckerle, M.C, Winge, D.R, Summers, M.F. | | Deposit date: | 1995-12-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the cysteine-rich intestinal protein, CRIP.
J.Mol.Biol., 257, 1996
|
|
1IMO
 
 | |
1IMQ
 
 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
1IMU
 
 | | Solution Structure of HI0257, a Ribosome Binding Protein | | Descriptor: | HYPOTHETICAL PROTEIN HI0257 | | Authors: | Parsons, L, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-11 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI0257, a bacterial ribosome binding protein.
Biochemistry, 40, 2001
|
|
1IN1
 
 | |
1IOG
 
 | | INSULIN MUTANT A3 GLY,(B1, B10, B16, B27)GLU, DES-B30, NMR, 19 STRUCTURES | | Descriptor: | PROTEIN (INSULIN PRECURSOR) | | Authors: | Olsen, H.B, Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1998-08-13 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The relationship between insulin bioactivity and structure in the NH2-terminal A-chain helix.
J.Mol.Biol., 284, 1998
|
|
1IOH
 
 | | INSULIN MUTANT A8 HIS,(B1, B10, B16, B27)GLU, DES-B30, NMR, 26 STRUCTURES | | Descriptor: | PROTEIN (INSULIN PRECURSOR) | | Authors: | Olsen, H.B, Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1998-08-13 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The relationship between insulin bioactivity and structure in the NH2-terminal A-chain helix.
J.Mol.Biol., 284, 1998
|
|
1IOJ
 
 | | HUMAN APOLIPOPROTEIN C-I, NMR, 18 STRUCTURES | | Descriptor: | APOC-I | | Authors: | Rozek, A, Sparrow, J.T, Weisgraber, K.H, Cushley, R.J. | | Deposit date: | 1998-05-12 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformation of human apolipoprotein C-I in a lipid-mimetic environment determined by CD and NMR spectroscopy.
Biochemistry, 38, 1999
|
|
1IOX
 
 | | NMR Structure of human Betacellulin-2 | | Descriptor: | Betacellulin | | Authors: | Miura, K, Doura, H, Aizawa, T, Tada, H, Seno, M, Yamada, H, Kawano, K. | | Deposit date: | 2001-04-18 | | Release date: | 2002-07-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of betacellulin, a new member of EGF-family ligands.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
1IP0
 
 | | NMR STRUCTURE OF HUMAN BETACELLULIN-2 | | Descriptor: | BETACELLULIN | | Authors: | Miura, K, Doura, H, Aizawa, T, Tada, H, Seno, M, Yamada, H, Kawano, K. | | Deposit date: | 2001-04-19 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of betacellulin, a new member of EGF-family ligands.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
1IP9
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
1IPG
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-05-14 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
1IQO
 
 | | Solution structure of MTH1880 from methanobacterium thermoautotrophicum | | Descriptor: | HYPOTHETICAL PROTEIN MTH1880 | | Authors: | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2001-07-23 | | Release date: | 2002-07-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
1IQS
 
 | | Minimized average structure of MTH1880 from Methanobacterium Thermoautotrophicum | | Descriptor: | MTH1880 | | Authors: | Lee, C.H, Shin, J, Bang, E, Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2001-07-29 | | Release date: | 2002-07-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel calcium binding protein, MTH1880, from Methanobacterium thermoautotrophicum.
Protein Sci., 13, 2004
|
|
1IR5
 
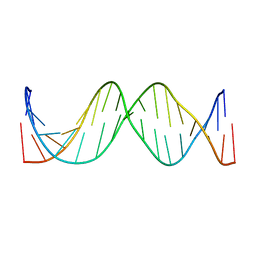 | | Solution Structure of the 17mer TF1 Binding Site | | Descriptor: | 5'-D(*CP*AP*CP*TP*AP*CP*AP*AP*AP*GP*AP*GP*TP*AP*GP*TP*G)-3', 5'-D(*CP*AP*CP*TP*AP*CP*TP*CP*TP*TP*TP*GP*TP*AP*GP*TP*G)-3' | | Authors: | Liu, W, Vu, H.M, Kearns, D.R. | | Deposit date: | 2001-09-07 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | 1H NMR studies of a 17-mer DNA duplex
ACTA BIOCHIM.BIOPHYS.SINICA, 1574, 2002
|
|
1IRH
 
 | | The Solution Structure of The Third Kunitz Domain of Tissue Factor Pathway Inhibitor | | Descriptor: | tissue factor pathway inhibitor | | Authors: | Mine, S, Yamazaki, T, Miyata, T, Hara, S, Kato, H. | | Deposit date: | 2001-10-02 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism for heparin-binding of the third Kunitz domain of human tissue factor pathway inhibitor.
Biochemistry, 41, 2002
|
|
1IRY
 
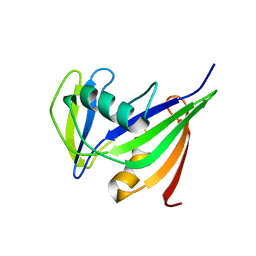 | | Solution structure of the hMTH1, a nucleotide pool sanitization enzyme | | Descriptor: | hMTH1 | | Authors: | Mishima, M, Itoh, N, Sakai, Y, Kamiya, H, Nakabeppu, Y, Shirakawa, M. | | Deposit date: | 2001-10-25 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of human MTH1, a Nudix family hydrolase that selectively degrades oxidized purine nucleoside triphosphates
J.Biol.Chem., 279, 2004
|
|
1IRZ
 
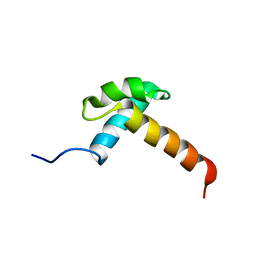 | |
1ISK
 
 | | 3-OXO-DELTA5-STEROID ISOMERASE, NMR, 20 STRUCTURES | | Descriptor: | 3-OXO-DELTA5-STEROID ISOMERASE | | Authors: | Wu, Z.R, Ebrahimian, S, Zawrotny, M.E, Thornburg, L.D, Perez-Alvarado, G.C, Brothers, P, Pollack, R.M, Summers, M.F. | | Deposit date: | 1997-03-12 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 3-oxo-delta5-steroid isomerase.
Science, 276, 1997
|
|
