5H3M
 
 | |
5H3N
 
 | |
5H7P
 
 | | NMR structure of the Vta1NTD-Did2(176-204) complex | | Descriptor: | Vacuolar protein sorting-associated protein VTA1, Vacuolar protein-sorting-associated protein 46 | | Authors: | Shen, J, Yang, Z, Wild, C.J. | | Deposit date: | 2016-11-20 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR studies on the interactions between yeast Vta1 and Did2 during the multivesicular bodies sorting pathway
Sci Rep, 6, 2016
|
|
5H7U
 
 | | NMR structure of eIF3 36-163 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit C | | Authors: | Nagata, T, Obayashi, E. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5.
Cell Rep, 18, 2017
|
|
5HOU
 
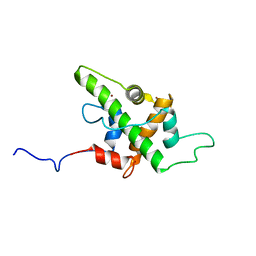 | | Solution Structure of p53TAD-TAZ1 | | Descriptor: | Cellular tumor antigen p53,CREB-binding protein fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HP0
 
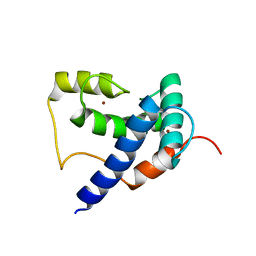 | | Solution Structure of TAZ2-p53AD2 | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HPD
 
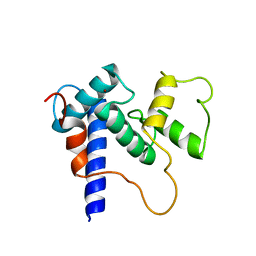 | | Solution Structure of TAZ2-p53TAD | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HQF
 
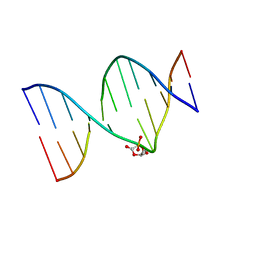 | | DNA duplex containing a ribonolactone lesion | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(RIB)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*GP*GP*AP*(8OG)P*CP*G)-3') | | Authors: | Zalesak, J, Constant, J.F, Jourdan, M. | | Deposit date: | 2016-01-21 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of DNA Featuring Clustered 2'-Deoxyribonolactone and 8-Oxoguanine Lesions.
Biochemistry, 55, 2016
|
|
5HQQ
 
 | | DNA duplex containing a ribonolactone lesion | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(RIB)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*(8OG)P*TP*GP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Zalesak, J, Constant, J.F, Jourdan, M. | | Deposit date: | 2016-01-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of DNA Featuring Clustered 2'-Deoxyribonolactone and 8-Oxoguanine Lesions.
Biochemistry, 55, 2016
|
|
5HUZ
 
 | | Solution structure of coiled coil domain of myosin binding subunit of myosin light chain phosphatase | | Descriptor: | Protein phosphatase 1 regulatory subunit 12A | | Authors: | Sharma, A.K, Birrane, G, Anklin, C, Rigby, A.C, Pollak, M, Alper, S.L. | | Deposit date: | 2016-01-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | To be published
To Be Published
|
|
5HV8
 
 | | Solution structure of an octanoyl- loaded acyl carrier protein domain from module MLSA2 of the mycolactone polyketide synthase. | | Descriptor: | S-[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] octanethioate, Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
5HVC
 
 | | Solution structure of the apo state of the acyl carrier protein from the MLSA2 subunit of the mycolactone polyketide synthase | | Descriptor: | Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
5I1R
 
 | | Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR and X-ray scattering | | Descriptor: | Nucleocapsid protein p7, ZINC ION | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Quantitative Characterization of Configurational Space Sampled by HIV-1 Nucleocapsid Using Solution NMR, X-ray Scattering and Protein Engineering.
Chemphyschem, 17, 2016
|
|
5I1X
 
 | |
5I22
 
 | | Amphiphysin SH3 in complex with Chikungunya virus nsP3 peptide | | Descriptor: | CHIKV nsP3 peptide, Myc box-dependent-interacting protein 1 | | Authors: | Tossavainen, H, Aitio, O, Hellman, M, Saksela, K, Permi, P. | | Deposit date: | 2016-02-04 | | Release date: | 2016-06-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the High Affinity Interaction between the Alphavirus Nonstructural Protein-3 (nsP3) and the SH3 Domain of Amphiphysin-2.
J.Biol.Chem., 291, 2016
|
|
5I2P
 
 | |
5I2V
 
 | |
5I65
 
 | |
5I8N
 
 | |
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
5IAZ
 
 | |
5ID3
 
 | | Solution structure of the pore-forming region of C. elegans Mitochondrial Calcium Uniporter (MCU) | | Descriptor: | Mitochondrial Calcium Uniporter | | Authors: | Oxenoid, K, Dong, Y, Cao, C, Cui, T, Sancak, Y, Markhard, A.L, Grabarek, Z, Kong, L, Liu, Z, Ouyang, B, Cong, Y, Mootha, V.K, Chou, J.J, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2016-02-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Architecture of the mitochondrial calcium uniporter.
Nature, 533, 2016
|
|
5ID5
 
 | |
5IE8
 
 | |
5IEB
 
 | |
