1H9C
 
 | | NMR structure of cysteinyl-phosphorylated enzyme IIB of the N,N'-diacetylchitobiose specific phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. | | Descriptor: | PTS SYSTEM, CHITOBIOSE-SPECIFIC IIB COMPONENT | | Authors: | Ab, E, Schuurman-Wolters, G.K, Nijlant, D, Dijkstra, K, Saier, M.H, Robillard, G.T, Scheek, R.M. | | Deposit date: | 2001-03-07 | | Release date: | 2001-05-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Cysteinyl-Phosphorylated Enzyme Iib of the N,N'-Diacetylchitobiose Specific Phosphoenolpyruvate-Dependentphosphotransferase System of Escherichia Coli
J.Mol.Biol., 308, 2001
|
|
1H9E
 
 | | LEM-LIKE DOMAIN OF HUMAN INNER NUCLEAR MEMBRANE PROTEIN LAP2 | | Descriptor: | LAMINA-ASSOCIATED POLYPEPTIDE 2 | | Authors: | Laguri, C, Gilquin, B, Wolff, N, Romi-Lebrun, R, Courchay, K, Callebaut, I, Worman, H.J, Zinn-Justin, S. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Lem Motif Common to Three Human Inner Nuclear Membrane Proteins
Structure, 9, 2001
|
|
1H9F
 
 | | LEM DOMAIN OF HUMAN INNER NUCLEAR MEMBRANE PROTEIN LAP2 | | Descriptor: | Lamina-associated polypeptide 2, isoform alpha | | Authors: | Laguri, C, Gilquin, B, Wolff, N, Romi-Lebrun, R, Courchay, K, Callebaut, I, Worman, H.J, Zinn-Justin, S. | | Deposit date: | 2001-03-09 | | Release date: | 2001-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Lem Motif Common to Three Human Inner Nuclear Membrane Proteins
Structure, 9, 2001
|
|
1HA6
 
 | | NMR Solution Structure of Murine CCL20/MIP-3a Chemokine | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN 3 ALPHA | | Authors: | Perez-Canadillas, J.M, Zaballos, A, Gutierrez, J, Varona, R, Roncal, F, Albar, J.P, Marquez, G, Bruix, M. | | Deposit date: | 2001-03-28 | | Release date: | 2001-08-22 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Murine Ccl20/Mip3-A, a Chemiokine that Specifically Chemoattracs Immature Dendritic Cells and Lymphocytes Through its Highly Specific Interaction with the Beta-Chemokine Receptor Ccr6
J.Biol.Chem., 276, 2001
|
|
1HA8
 
 | |
1HA9
 
 | | SOLUTION STRUCTURE OF THE SQUASH TRYPSIN INHIBITOR MCoTI-II, NMR, 30 STRUCTURES. | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Heitz, A, Hernandez, J.-F, Gagnon, J, Hong, T.T, Pham, T.T.C, Nguyen, T.M, Le-Nguyen, D, Chiche, L. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Squash Trypsin Inhibitor Mcoti-II. A New Family for Cyclic Knottins
Biochemistry, 40, 2001
|
|
1HAA
 
 | | A beta-Hairpin Structure in a 13-mer Peptide that Binds a-Bungarotoxin with High Affinity and Neutralizes its Toxicity | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE | | Authors: | Scherf, T, Kasher, R, Balass, M, Fridkin, M, Fuchs, S, Katchalski-Katzir, E. | | Deposit date: | 2001-04-05 | | Release date: | 2001-05-25 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A Beta-Hairpin Structure in a 13-mer Peptide that Binds Alpha-Bungarotoxin with High Affinity and Neutralizes its Toxicity
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HAJ
 
 | | A beta-Hairpin Structure in a 13-mer Peptide that Binds a-Bungarotoxin with High Affinity and Neutralizes its Toxicity | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE | | Authors: | Scherf, T, Kasher, R, Balass, M, Fridkin, M, Fuchs, S, Katchalski-Katzir, E. | | Deposit date: | 2001-04-05 | | Release date: | 2001-05-25 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A Beta-Hairpin Structure in a 13-mer Peptide that Binds Alpha-Bungarotoxin with High Affinity and Neutralizes its Toxicity
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HBW
 
 | | Solution nmr structure of the dimerization domain of the yeast transcriptional activator Gal4 (residues 50-106) | | Descriptor: | REGULATORY PROTEIN GAL4 | | Authors: | Hidalgo, P, Ansari, A.Z, Schmidt, P, Hare, B, Simkovic, N, Farrell, S, Shin, E.J, Ptashne, M, Wagner, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Recruitment of the Transcriptional Machinery Through Gal11P: Structure and Interactions of the GAL4 Dimerization Domain
Genes Dev., 15, 2001
|
|
1HD4
 
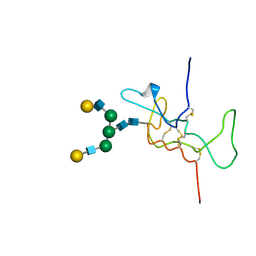 | | SOLUTION STRUCTURE OF THE A-SUBUNIT OF HUMAN CHORIONIC GONADOTROPIN [MODELED WITH DIANTENNARY GLYCAN AT ASN78] | | Descriptor: | CHORIONIC GONADOTROPIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, van Kuik, J.A, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Effects of the N-Linked Glycans on the 3D Structure of the Free A-Subunit of Human Chorionic Gonadotropin
Biochemistry, 39, 2000
|
|
1HD6
 
 | | PHEROMONE ER-22, NMR | | Descriptor: | PHEROMONE ER-22 | | Authors: | Luginbuhl, P, Liu, A, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 2000-11-09 | | Release date: | 2000-12-10 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Pheromone Er-22 from Euplotes Raikovi
J.Biomol.NMR, 19, 2001
|
|
1HD9
 
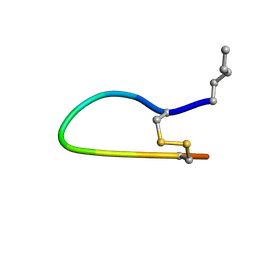 | | The Bowman-Birk Inhibitor Reactive Site Loop Sequence Represents an Independent Structural Beta-Hairpin Motif | | Descriptor: | BOWMAN-BIRK INHIBITOR DERIVED PEPTIDE | | Authors: | Brauer, A.B.E, Kelly, G, McBride, J.D, Cooke, R.M, Matthews, S.J, Leatherbarrow, R.J. | | Deposit date: | 2000-11-13 | | Release date: | 2001-03-29 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The Bowman-Birk Inhibitor Reactive Site Loop Sequence Represents an Independent Structural Beta-Hairpin Motif
J.Mol.Biol., 306, 2001
|
|
1HDJ
 
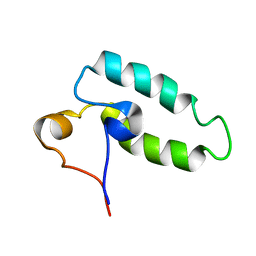 | | HUMAN HSP40 (HDJ-1), NMR | | Descriptor: | HUMAN HSP40 | | Authors: | Qian, Y.Q, Patel, D, Hartl, F.-U, Mccoll, D.J. | | Deposit date: | 1996-05-09 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain.
J.Mol.Biol., 260, 1996
|
|
1HDL
 
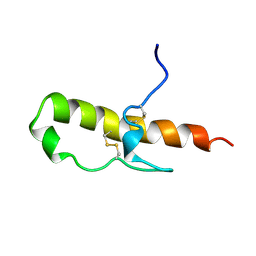 | | LEKTI domain one | | Descriptor: | SERINE PROTEINASE INHIBITOR LEKTI | | Authors: | Lauber, T, Roesch, P, Marx, U.C. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Homologous Proteins with Different Folds: The Three-Dimensional Structures of Domains 1 and 6 of the Multiple Kazal-Type Inhibitor Lekti
J.Mol.Biol., 328, 2003
|
|
1HEH
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
1HEJ
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
1HF9
 
 | | C-Terminal Coiled-Coil Domain from Bovine IF1 | | Descriptor: | ATPASE INHIBITOR (MITOCHONDRIAL) | | Authors: | Gordon-Smith, D.J, Carbajo, R.J, Yang, J.-C, Videler, H, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2000-11-30 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a C-terminal coiled-coil domain from bovine IF(1): the inhibitor protein of F(1) ATPase.
J. Mol. Biol., 308, 2001
|
|
1HFF
 
 | | NMR solution structures of the vMIP-II 1-10 peptide from Kaposi's sarcoma-associated herpesvirus. | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-01 | | Release date: | 2000-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
1HFG
 
 | | NMR solution structure of vMIP-II 1-71 from Kaposi's sarcoma-associated herpesvirus (minimized average structure). | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.-H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-01 | | Release date: | 2001-01-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
1HFN
 
 | | NMR solution structures of vMIP-II 1-71 from Kaposi's sarcoma-associated herpesvirus. | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.-H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-07 | | Release date: | 2001-01-07 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
1HG9
 
 | | Solution structure of DNA:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*TP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-13 | | Release date: | 2002-01-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity.
Chemistry, 6, 2000
|
|
1HI7
 
 | |
1HIC
 
 | |
1HIQ
 
 | |
1HIS
 
 | |
