1YYX
 
 | |
1YZA
 
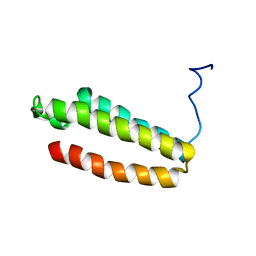 | | The solution structure of a redesigned apocytochrome B562 (Rd-apocyt b562) with the N-terminal helix unfolded | | Descriptor: | Redesigned apo-cytochrome b562 | | Authors: | Feng, H, Takei, T, Lipsitz, R, Tjandra, N, Bai, Y, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-02-28 | | Release date: | 2005-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Specific non-native hydrophobic interactions in a hidden folding intermediate: implication for protein folding
Biochemistry, 42, 2003
|
|
1YZC
 
 | |
1YZS
 
 | |
1Z00
 
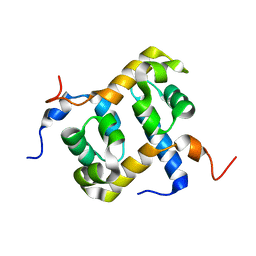 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
1Z09
 
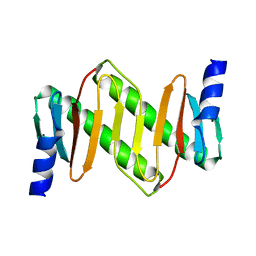 | | Solution structure of km23 | | Descriptor: | Dynein light chain 2A, cytoplasmic | | Authors: | Ilangovan, U, Ding, W, Mulder, K, Hinck, A.P, Zuniga, J, Trbovich, J.A, Demeler, B, Groppe, J.C. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Homodimeric Dynein Light Chain km23.
J.Mol.Biol., 352 (2), 2005
|
|
1Z0R
 
 | | Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transcription-State Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Bobay, B.G, Andreeva, A, Mueller, G.A, Cavanagh, J, Murzin, A.G. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins.
Febs Lett., 579, 2005
|
|
1Z1D
 
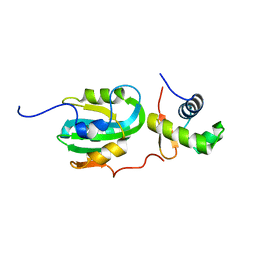 | | Structural Model for the interaction between RPA32 C-terminal domain and SV40 T antigen origin binding domain. | | Descriptor: | Large T antigen, Replication protein A 32 kDa subunit | | Authors: | Arunkumar, A.I, Klimovich, V, Jiang, X, Ott, R.D, Mizoue, L, Fanning, E, Chazin, W.J. | | Deposit date: | 2005-03-03 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1Z1M
 
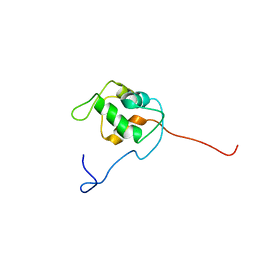 | | NMR structure of unliganded MDM2 | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Uhrinova, S, Uhrin, D, Powers, H, Watt, K, Zheleva, D, Fischer, P, McInnes, C, Barlow, P.N. | | Deposit date: | 2005-03-04 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Free MDM2 N-terminal Domain Reveals Conformational Adjustments that Accompany p53-binding
J.Mol.Biol., 350, 2005
|
|
1Z1V
 
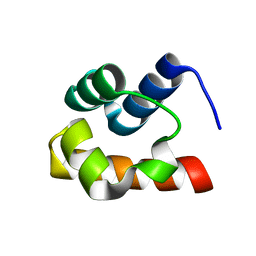 | | NMR structure of the Saccharomyces cerevisiae Ste50 SAM domain | | Descriptor: | STE50 protein | | Authors: | Kwan, J.J, Warner, N, Maini, J, Pawson, T, Donaldson, L.W. | | Deposit date: | 2005-03-06 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Saccharomyces cerevisiae Ste50 binds the MAPKKK Ste11 through a head-to-tail SAM domain interaction.
J.Mol.Biol., 356, 2006
|
|
1Z1Z
 
 | |
1Z23
 
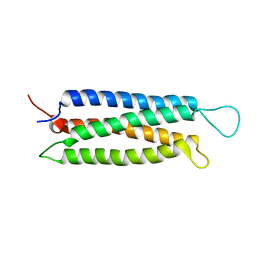 | | The serine-rich domain from Crk-associated substrate (p130Cas) | | Descriptor: | CRK-associated substrate | | Authors: | Briknarova, K, Nasertorabi, F, Havert, M.L, Eggleston, E, Hoyt, D.W, Li, C, Olson, A.J, Vuori, K, Ely, K.R. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The serine-rich domain from Crk-associated substrate (p130cas) is a four-helix bundle.
J.Biol.Chem., 280, 2005
|
|
1Z2D
 
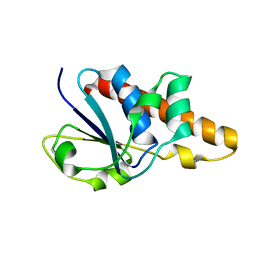 | | Solution Structure of Bacillus subtilis ArsC in reduced state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
1Z2E
 
 | | Solution Structure of Bacillus subtilis ArsC in oxidized state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
1Z2G
 
 | | Solution structure of apo, oxidized yeast Cox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Arnesano, F, Balatri, E, Banci, L, Bertini, I, Winge, D.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-03-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding
STRUCTURE, 13, 2005
|
|
1Z2J
 
 | |
1Z2K
 
 | | NMR structure of the D1 domain of the Natural Killer Cell Receptor, 2B4 | | Descriptor: | Natural killer cell receptor 2B4 | | Authors: | Ames, J.B, Vyas, V, Lusin, J.D, Mariuzza, R. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Natural Killer Cell Receptor 2B4:
Implications for Ligand Recognition
Biochemistry, 44, 2005
|
|
1Z2Q
 
 | |
1Z2T
 
 | | NMR structure study of anchor peptide Ser65-Leu87 of enzyme acholeplasma laidlawii Monoglycosyldiacyl Glycerol Synthase (alMGS) in DHPC micelles | | Descriptor: | Anchor peptide Ser65-Leu87 of alMGS | | Authors: | Lind, J, Barany-Wallje, E, Ramo, T, Wieslander, A, Maler, L. | | Deposit date: | 2005-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, position of and membrane-interaction of a putative membrane-anchoring domain of alMGS
To be Published
|
|
1Z30
 
 | | NMR structure of the apical part of stemloop D from cloverleaf 1 of bovine enterovirus 1 RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*CP*GP*UP*UP*AP*GP*AP*AP*CP*GP*UP*C)-3' | | Authors: | Ihle, Y, Ohlenschlager, O, Duchardt, E, Ramachandran, R, Gorlach, M. | | Deposit date: | 2005-03-10 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA
Nucleic Acids Res., 33, 2005
|
|
1Z31
 
 | |
1Z3K
 
 | |
1Z3R
 
 | | Solution structure of the Omsk Hemhorraghic Fever Envelope Protein Domain III | | Descriptor: | polyprotein | | Authors: | Volk, D.E, Chavez, L, Beasley, D.W, Barrett, A.D, Gorenstein, D.G. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the envelope protein domain III of Omsk hemorrhagic fever virus.
Virology, 351, 2006
|
|
1Z4H
 
 | | The response regulator TorI belongs to a new family of atypical excisionase | | Descriptor: | Tor inhibition protein | | Authors: | Elantak, L, Ansaldi, M, Guerlesquin, F, Mejean, V, Morelli, X. | | Deposit date: | 2005-03-16 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and genetic analyses reveal a key role in prophage excision for the TorI response regulator inhibitor
J.Biol.Chem., 280, 2005
|
|
1Z5F
 
 | | Solution Structure of the Cytotoxic RC-RNase 3 with a Pyroglutamate Residue at the N-terminus | | Descriptor: | RC-RNase 3 | | Authors: | Lou, Y.C, Huang, Y.C, Pan, Y.R, Chen, C, Liao, Y.D. | | Deposit date: | 2005-03-18 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3
J.Mol.Biol., 355, 2006
|
|
