1YSV
 
 | |
1YSY
 
 | | NMR Structure of the nonstructural Protein 7 (nsP7) from the SARS CoronaVirus | | Descriptor: | Replicase polyprotein 1ab (pp1ab) (ORF1AB) | | Authors: | Peti, W, Herrmann, T, Johnson, M.A, Kuhn, P, Stevens, R.C, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-02-09 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7.
J.Virol., 79, 2005
|
|
1YT6
 
 | | NMR structure of peptide SD | | Descriptor: | peptide SD | | Authors: | Murata, T, Hemmi, H, Nakamura, S, Shimizu, K, Suzuki, Y, Yamaguchi, I. | | Deposit date: | 2005-02-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure, epitope mapping, and docking simulation of a gibberellin mimic peptide as a peptidyl mimotope for a hydrophobic ligand.
Febs J., 272, 2005
|
|
1YTP
 
 | |
1YTR
 
 | | NMR structure of plantaricin a in dpc micelles, 20 structures | | Descriptor: | Bacteriocin plantaricin A | | Authors: | Kristiansen, P.E, Fimland, G, Mantzilas, D, Nissen-Meyer, J. | | Deposit date: | 2005-02-11 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and mode of action of the membrane-permeabilizing antimicrobial peptide pheromone plantaricin A
J.Biol.Chem., 280, 2005
|
|
1YUI
 
 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1996-12-31 | | Release date: | 1997-12-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
1YUS
 
 | | Solution structure of apo-S100A13 | | Descriptor: | S100 calcium binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YUU
 
 | | Solution structure of Calcium-S100A13 | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YV8
 
 | |
1YVA
 
 | | NMR solution structure of crambin in DPC micelles | | Descriptor: | Crambin | | Authors: | Ahn, H.-C, Markley, J.L. | | Deposit date: | 2005-02-15 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of the Water-Insoluble Protein Crambin in Dodecylphosphocholine Micelles and Its Minimal Solvent-Exposed Surface
J.Am.Chem.Soc., 128, 2006
|
|
1YVC
 
 | | Solution structure of the conserved protein from the gene locus MMP0076 of Methanococcus maripaludis. Northeast Structural Genomics target MrR5. | | Descriptor: | MrR5 | | Authors: | Northeast Structural Genomics Consortium (NESG), Rossi, P, Aramini, J.M, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T. | | Deposit date: | 2005-02-15 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: |
To be Published
|
|
1YWI
 
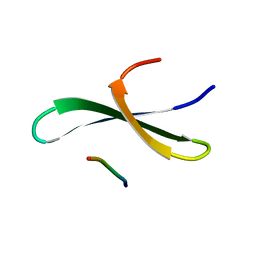 | | Structure of the FBP11WW1 domain complexed to the peptide APPTPPPLPP | | Descriptor: | Formin, Formin-binding protein 3 | | Authors: | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
1YWJ
 
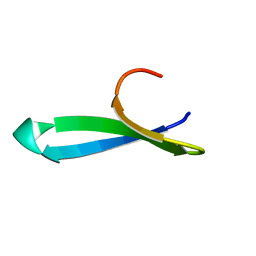 | | Structure of the FBP11WW1 domain | | Descriptor: | Formin-binding protein 3 | | Authors: | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
1YWL
 
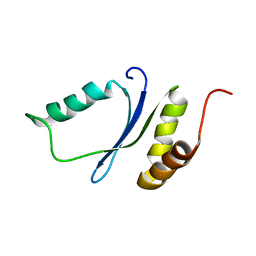 | |
1YWS
 
 | |
1YWU
 
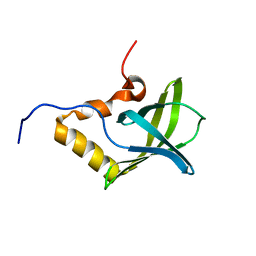 | | Solution NMR structure of Pseudomonas Aeruginosa protein PA4608. Northeast Structural Genomics target PaT7 | | Descriptor: | hypothetical protein PA4608 | | Authors: | Ramelot, T.A, Yee, A.A, Cort, J.R, Semesi, A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein.
Proteins, 66, 2007
|
|
1YWW
 
 | |
1YWX
 
 | | Solution Structure of Methanococcus maripaludis Protein MMP0443: The Northeast Structural Genomics Consortium Target MrR16 | | Descriptor: | 30S ribosomal protein S24e | | Authors: | Yang, C.S, Acton, T, Shen, Y, Ma, L, Liu, G, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methanococcus maripaludis Protein MMP0443: The Northeast Structural Genomics Consortium Target MrR16
To be Published
|
|
1YWY
 
 | | Solution Structure of Pseudomonas aeruginosa Protein PA2021. The Northeast Structural Genomics Consortium Target Pat85. | | Descriptor: | hypothetical protein PA2021 | | Authors: | Lin, Y.C, Liu, G, Shen, Y, Yee, A, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Pseudomonas aeruginosa Protein PA2021.
To be Published
|
|
1YX0
 
 | | Solution Structure of Bacillus subtilis Protein ysnE: The Northeast Structural Genomics Consortium Target SR220 | | Descriptor: | hypothetical protein ysnE | | Authors: | Liu, G, Ma, L, Shen, Y, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Bacillus subtilis Protein ysnE
To be Published
|
|
1YX3
 
 | | NMR structure of Allochromatium vinosum DsrC: Northeast Structural Genomics Consortium target OP4 | | Descriptor: | hypothetical protein DsrC | | Authors: | Cort, J.R, Dahl, C, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation.
J.Mol.Biol., 382, 2008
|
|
1YXR
 
 | | NMR Structure of VPS4A MIT Domain | | Descriptor: | vacuolar protein sorting factor 4A | | Authors: | Scott, J.A, Gaspar, J, Stuchell, M, Alam, S, Skalicky, J, Sundquist, W.I. | | Deposit date: | 2005-02-22 | | Release date: | 2005-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of VPS4A MIT Domain
To be Published
|
|
1YYB
 
 | |
1YYC
 
 | |
1YYJ
 
 | | The NMR solution structure of a redesigned apocytochrome b562:Rd-apocyt b562 | | Descriptor: | redesigned apocytochrome B562 | | Authors: | Feng, H, Takei, J, Lipsitz, R, Tjandra, N, Bai, Y, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-02-25 | | Release date: | 2005-08-25 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Specific non-native hydrophobic interactions in a hidden folding intermediate: implications for protein folding
Biochemistry, 42, 2003
|
|
