8ZUG
 
 | |
9ATN
 
 | |
9AZI
 
 | |
9BAF
 
 | | Solution NMR structure of conofurin-Delta | | Descriptor: | Alpha-conotoxin LvIA | | Authors: | Harvey, P.J, Craik, D.J, Hone, A.J, McIntosh, J.M. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Novel Peptide Derivatives of the Severe Acute Respiratory Syndrome-Coronavirus-2 Spike-Protein that Potently Inhibit Nicotinic Acetylcholine Receptors.
J.Med.Chem., 67, 2024
|
|
9BB1
 
 | |
9BB2
 
 | |
9BB3
 
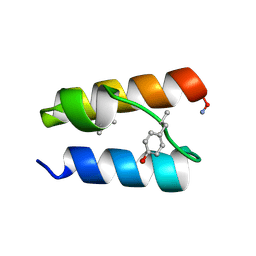 | |
9BB4
 
 | |
9BB5
 
 | |
9BB6
 
 | |
9BB7
 
 | |
9BD0
 
 | |
9BFL
 
 | |
9BHN
 
 | |
9BV0
 
 | |
9BVS
 
 | | NMR structure of TLP-1 in solution | | Descriptor: | Temporin-1Tl | | Authors: | Jia, R, McShan, A.C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Design and Development of Temporin L Analogues to Inhibit the Main Protease of SARS-CoV-2
To Be Published
|
|
9BVU
 
 | | NMR structure of TLP-2 in solution | | Descriptor: | Temporin-1Tl | | Authors: | Jia, R, McShan, A.C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Design and Development of Temporin L Analogues to Inhibit the Main Protease of SARS-CoV-2
To Be Published
|
|
9BVV
 
 | | NMR structure of TLP-3 in solution | | Descriptor: | Temporin-1Tl | | Authors: | Jia, R, McShan, A.C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Design and Development of Temporin L Analogues to Inhibit the Main Protease of SARS-CoV-2
To Be Published
|
|
9C5E
 
 | | Covalent Complex Between Parkin Catalytic (Rcat) Domain and Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ZINC ION | | Authors: | Connelly, E.M, Rintala-Dempsey, A.C, Gundogdu, M, Freeman, E.A, Koszela, J, Aguirre, J.D, Zhu, G, Kamarainen, O, Tadayon, R, Walden, H, Shaw, G.S. | | Deposit date: | 2024-06-06 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Capturing the catalytic intermediates of parkin ubiquitination.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9CE9
 
 | |
9CEQ
 
 | |
9CJA
 
 | |
9COJ
 
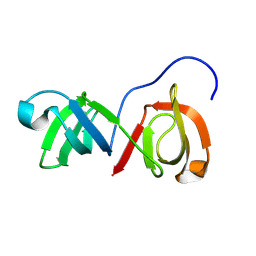 | | SH3-like tandem domain of human KIN protein | | Descriptor: | DNA/RNA-binding protein KIN17 | | Authors: | de Lourenco, I.O, Fossey, M.A, Souza, F.P, Seixas, F.A.V, Fernandez, M.A, Almeida, F.C.L, Caruso, I.P. | | Deposit date: | 2024-07-16 | | Release date: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3-like tandem domain of human KIN protein and its interaction with RNA homo-oligonucleotides
To Be Published
|
|
9CPD
 
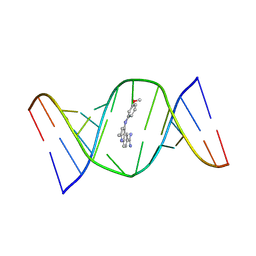 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[(3-methoxyphenyl)amino]-2-methylquinoline-6-carboximidamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
9CPG
 
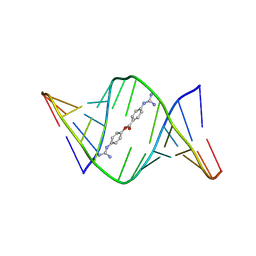 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-carbamimidamidophenyl 4-carbamimidamidobenzoate, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
