1K8H
 
 | |
1K8J
 
 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8M
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8O
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
1K8V
 
 | |
1K91
 
 | | Solution Structure of Calreticulin P-domain subdomain (residues 221-256) | | Descriptor: | CALRETICULIN | | Authors: | Ellgaard, L, Bettendorff, P, Braun, D, Herrmann, T, Fiorito, F, Guntert, P, Helenius, A, Wuthrich, K. | | Deposit date: | 2001-10-26 | | Release date: | 2002-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of 36 and 73-residue Fragments of the Calreticulin P-domain
J.Mol.Biol., 322, 2002
|
|
1K9C
 
 | | Solution Structure of Calreticulin P-domain subdomain (residues 189-261) | | Descriptor: | CALRETICULIN | | Authors: | Ellgaard, L, Bettendorff, P, Braun, D, Herrmann, T, Fiorito, F, Guntert, P, Helenius, A, Wuthrich, K. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of 36 and 73-residue Fragments of the Calreticulin P-domain
J.Mol.Biol., 322, 2002
|
|
1K9Q
 
 | | YAP65 WW domain complexed to N-(n-octyl)-GPPPY-NH2 | | Descriptor: | 65 kDa Yes-associated protein, N-OCTANE, WW domain binding protein-1 | | Authors: | Pires, J.R, Taha-Nejad, F, Toepert, F, Ast, T, Hoffmuller, U, Schneider-Mergener, J, Kuhne, R, Macias, M.J, Oschkinat, H. | | Deposit date: | 2001-10-30 | | Release date: | 2001-12-28 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope.
J.Mol.Biol., 314, 2001
|
|
1K9R
 
 | | YAP65 WW domain complexed to Acetyl-PLPPY | | Descriptor: | 65 kDa Yes-associated protein, WW domain binding protein-1 | | Authors: | Pires, J.R, Taha-Nejad, F, Toepert, F, Ast, T, Hoffmuller, U, Schneider-Mergener, J, Kuhne, R, Macias, M.J, Oschkinat, H. | | Deposit date: | 2001-10-30 | | Release date: | 2001-12-28 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope.
J.Mol.Biol., 314, 2001
|
|
1KA6
 
 | | SAP/SH2D1A bound to peptide n-pY | | Descriptor: | SH2 DOMAIN PROTEIN 1A, peptide n-pY | | Authors: | Hwang, P.M, Li, C, Morra, M, Lillywhite, J, Gertler, F, Terhorst, C, Kay, L.E, Pawson, T, Forman-Kay, J, Li, S.-C. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A "three-pronged" binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome.
EMBO J., 21, 2002
|
|
1KA7
 
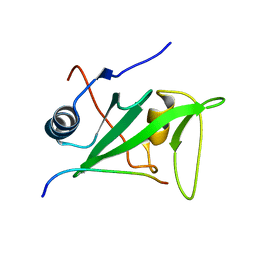 | | SAP/SH2D1A bound to peptide n-Y-c | | Descriptor: | SH2 DOMAIN PROTEIN 1A, peptide n-Y-c | | Authors: | Hwang, P.M, Li, C, Morra, M, Lillywhite, J, Gertler, F, Terhorst, C, Kay, L.E, Pawson, T, Forman-Kay, J, Li, S.-C. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A "three-pronged" binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome.
EMBO J., 21, 2002
|
|
1KAJ
 
 | |
1KAL
 
 | |
1KB1
 
 | |
1KB7
 
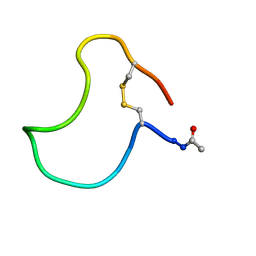 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | KB7 PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1KBH
 
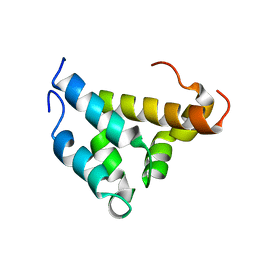 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
1KBM
 
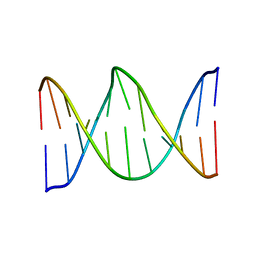 | |
1KBS
 
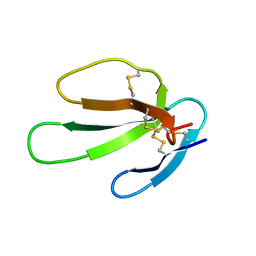 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 1 STRUCTURE | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
1KC4
 
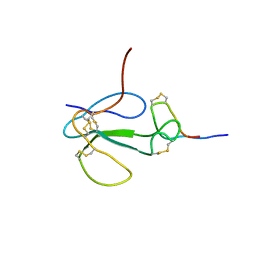 | | NMR Structural Analysis of the Complex Formed Between alpha-Bungarotoxin and the Principal alpha-Neurotoxin Binding Sequence on the alpha7 Subunit of a Neuronal Nicotinic Acetylcholine Receptor | | Descriptor: | alpha-bungarotoxin, neuronal acetylcholine receptor protein, alpha-7 chain | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-11-07 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
1KDF
 
 | | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Sonnichsen, F.D, Deluca, C.I, Davies, P.L, Sykes, B.D. | | Deposit date: | 1996-07-08 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction.
Structure, 4, 1996
|
|
1KDL
 
 | |
1KDX
 
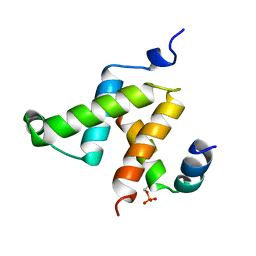 | | KIX DOMAIN OF MOUSE CBP (CREB BINDING PROTEIN) IN COMPLEX WITH PHOSPHORYLATED KINASE INDUCIBLE DOMAIN (PKID) OF RAT CREB (CYCLIC AMP RESPONSE ELEMENT BINDING PROTEIN), NMR 17 STRUCTURES | | Descriptor: | CBP, CREB | | Authors: | Radhakrishnan, I, Perez-Alvarado, G.C, Dyson, H.J, Wright, P.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-11-25 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions.
Cell(Cambridge,Mass.), 91, 1997
|
|
1KFT
 
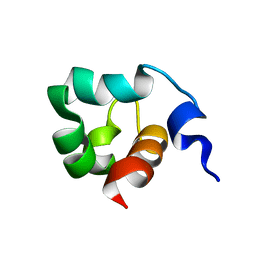 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
