6PRI
 
 | |
6PRJ
 
 | |
6PRP
 
 | |
6PRQ
 
 | |
6PSI
 
 | |
6PTS
 
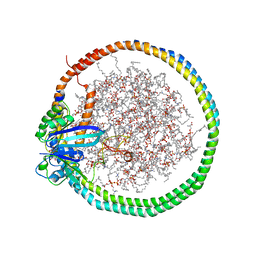 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state A) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PTW
 
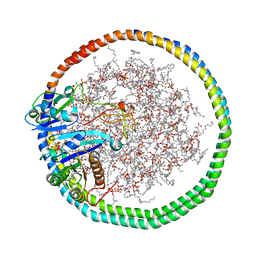 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state B) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PV0
 
 | |
6PV1
 
 | |
6PV2
 
 | |
6PV3
 
 | |
6PVR
 
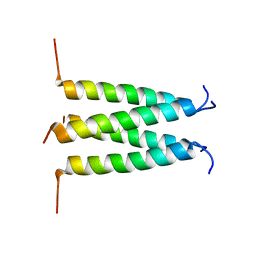 | | Influenza B M2 Proton Channel in the Closed State - SSNMR Structure at pH 7.5 | | Descriptor: | BM2 protein | | Authors: | Mandala, V.S, Loftis, A.R, Shcherbakov, A.S, Pentelute, B.L, Hong, M. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PVT
 
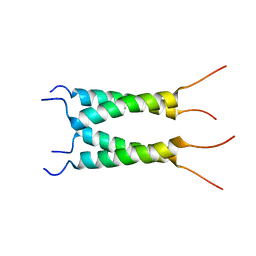 | | Influenza B M2 Proton Channel in the Open State - SSNMR Structure at pH 4.5 | | Descriptor: | BM2 protein | | Authors: | Mandala, V.S, Loftis, A.R, Shcherbakov, A.S, Pentelute, B.L, Hong, M. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PX7
 
 | |
6PX8
 
 | |
6Q08
 
 | |
6Q1X
 
 | | Lasso peptide pandonodin | | Descriptor: | Pandonodin | | Authors: | Link, A.J, Cheung-Lee, W.L. | | Deposit date: | 2019-08-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pandonodin: A Proteobacterial Lasso Peptide with an Exceptionally Long C-Terminal Tail.
Acs Chem.Biol., 14, 2019
|
|
6Q2I
 
 | |
6Q2Z
 
 | | NMR solution structure of the HVO_2922 protein from Haloferax volcanii | | Descriptor: | UPF0339 family protein | | Authors: | Kubatova, N, Jonker, H.R.A, Saxena, K, Richter, C, Marchfelder, A, Schwalbe, H. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Small Protein HVO_2922 from Haloferax volcanii.
Chembiochem, 21, 2020
|
|
6Q44
 
 | | Est3 telomerase subunit in the yeast Hansenula polymorpha | | Descriptor: | Uncharacterized protein | | Authors: | Mantsyzov, A.B, Mariasina, S.S, Petrova, O.A, Efimov, S.V, Dontsova, O.A, Polshakov, V.I. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights into the structure and function of Est3 from the Hansenula polymorpha telomerase.
Sci Rep, 10, 2020
|
|
6Q5Z
 
 | | H-Vc7.2, H-superfamily conotoxin | | Descriptor: | Conotoxin Vc7.2 | | Authors: | Nielsen, L.D, Foged, M.M, Teilum, K, Ellgaard, L. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of an H-superfamily conotoxin reveals a granulin fold arising from a common ICK cysteine framework.
J.Biol.Chem., 294, 2019
|
|
6Q6E
 
 | | Structural and functional insights into the condensin ATPase cycle | | Descriptor: | Condensin complex subunit 2,Structural maintenance of chromosomes protein,Structural maintenance of chromosomes protein | | Authors: | Simon, B, Hassler, M, Haering, C.H, Hennig, J. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
6QAM
 
 | |
6QAN
 
 | |
6QAX
 
 | | P31-43 | | Descriptor: | LEU-GLY-GLN-GLN-GLN-PRO-PHE-PRO-PRO-GLN-GLN-PRO-TYR | | Authors: | Calvanese, L, D'Auria, G, Falcigno, F. | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural insights on P31-43, a gliadin peptide able to promote an innate but not an adaptive response in celiac disease.
J.Pept.Sci., 25, 2019
|
|
