5XE4
 
 | |
5XEE
 
 | |
5XEK
 
 | |
5XER
 
 | | TK9 NMR structure in DPC micelle | | Descriptor: | THR-VAL-TYR-VAL-TYR-SER-ARG-VAL-LYS | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights of a self-assembling 9-residue peptide from the C-terminal tail of the SARS corona virus E-protein in DPC and SDS micelles: A combined high and low resolution spectroscopic study.
Biochim Biophys Acta Biomembr, 1860, 2018
|
|
5XES
 
 | | TK9 NMR structure in SDS micelle | | Descriptor: | THR-VAL-TYR-VAL-TYR-SER-ARG-VAL-LYS | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights of a self-assembling 9-residue peptide from the C-terminal tail of the SARS corona virus E-protein in DPC and SDS micelles: A combined high and low resolution spectroscopic study.
Biochim Biophys Acta Biomembr, 1860, 2018
|
|
5XF0
 
 | |
5XHT
 
 | | The PHD finger of human Kiaa1045 protein | | Descriptor: | PHD finger protein 24, ZINC ION | | Authors: | Miyamoto, K. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD finger from the human KIAA1045 protein
Protein Sci., 27, 2018
|
|
5XI1
 
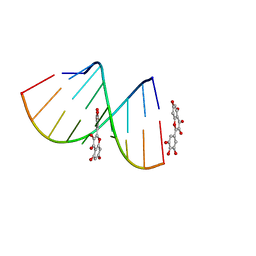 | | Structural Insight of Flavonoids binding to CAG repeat RNA that causes Huntington's Disease (HD) and Spinocerebellar Ataxia (SCAs) | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, RNA (5'-R(P*CP*CP*GP*CP*AP*GP*CP*GP*G)-3') | | Authors: | Tawani, A, Mishra, S.K, Khan, E, Kumar, A. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Myricetin Reduces Toxic Level of CAG Repeats RNA in Huntington's Disease (HD) and Spino Cerebellar Ataxia (SCAs).
ACS Chem. Biol., 13, 2018
|
|
5XI9
 
 | |
5XIR
 
 | |
5XIV
 
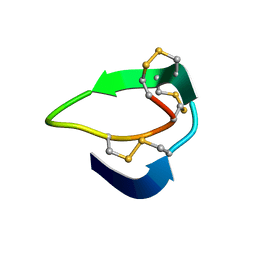 | | Beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba | | Descriptor: | beta-ginkgotide, beta-gB1 | | Authors: | Wong, K.H, Tan, W.L, Xiao, T, Tam, J.P. | | Deposit date: | 2017-04-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba.
Sci Rep, 7, 2017
|
|
5XJK
 
 | |
5XK4
 
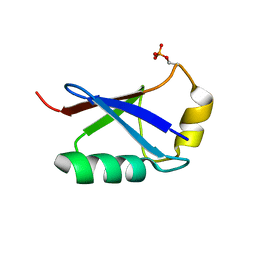 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XK5
 
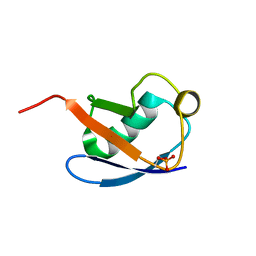 | | Relaxed state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Xu, D, Zhou, G, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XM4
 
 | |
5XME
 
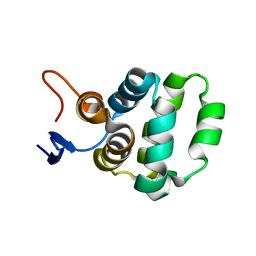 | | Solution structure of C-terminal domain of TRADD | | Descriptor: | Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2017-05-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily.
Sci Rep, 7, 2017
|
|
5XN4
 
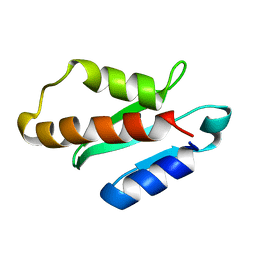 | | Anti-CRISPR protein AcrIIA4 | | Descriptor: | Anti-CRISPR AcrIIA4 | | Authors: | Suh, J.-Y, Kim, I. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor.
Sci Rep, 8, 2018
|
|
5XND
 
 | |
5XNG
 
 | | EFK17A structure in Microgel MAA60 | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2017-05-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
5XO3
 
 | |
5XO4
 
 | |
5XO5
 
 | |
5XO9
 
 | |
5XOA
 
 | |
5XOK
 
 | |
