2N90
 
 | |
2N91
 
 | | A key amino acid in the control of different functional behavior within the triheme cytochrome family from Geobacter sulfurreducens | | Descriptor: | Cytochrome C, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dantas, J.M, Simoes, T, Bruix, M, Salgueiro, C.A. | | Deposit date: | 2015-11-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Unveiling the Structural Basis That Regulates the Energy Transduction Properties within a Family of Triheme Cytochromes from Geobacter sulfurreducens.
J.Phys.Chem.B, 120, 2016
|
|
2N92
 
 | | Solution structure of cecropin P1 with LPS | | Descriptor: | Cecropin-P1 | | Authors: | Baek, M, Kamiya, M, Kushibiki, T, Nakazumi, T, Tomisawa, S, Abe, C, Kumaki, Y, Kikukawa, T, Demura, M, Kawano, K, Aizawa, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Lipopolysaccharide bound structure of antimicrobial peptide cecropin P1 by NMR spectroscopy
To be Published
|
|
2N93
 
 | | Solution structure of lcFABP | | Descriptor: | Fatty acid-binding protein | | Authors: | Tseng, K, Lyu, P. | | Deposit date: | 2015-11-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments of light organ-associated fatty acid-binding protein of Taiwanese fireflies.
Biomol NMR Assign, 10, 2016
|
|
2N94
 
 | |
2N95
 
 | |
2N96
 
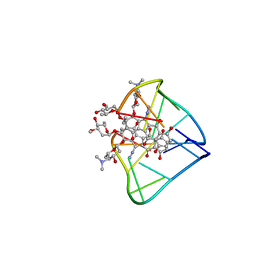 | | An unexpected mode of small molecule DNA binding provides the structural basis for DNA cleavage by the potent antiproliferative agent (-)-lomaiviticin A | | Descriptor: | (1R,1'R,2S,2'S,3R,3'R,5aR,10aR,11a'S)-2'-[(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)oxy]-2,2'-diethyl-11,11 '-dihydrazinyl-6,6',9,9'-tetrahydroxy-4,4',5,5',10,10'-hexaoxo-1,1'-bis{[2,4,6-trideoxy-4-(dimethylamino)-beta-L-arabino -hexopyranosyl]oxy}[2,2',3,3',4,4',5,5',5a,8,10,10',10a,11a'-tetradecahydro-1H,1'H-[3,3'-bibenzo[b]fluorene]]-2-yl 2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranoside, DNA (5'-D(*GP*CP*TP*AP*TP*AP*GP*C)-3') | | Authors: | Woo, C.M, Li, Z, Paulson, E, Herzon, S.B. | | Deposit date: | 2015-11-07 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA cleavage by the potent antiproliferative agent (-)-lomaiviticin A.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N97
 
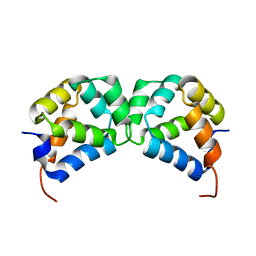 | | DD homodimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C.F. | | Deposit date: | 2015-11-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of death domain signaling in the p75 neurotrophin receptor
Elife, 4, 2015
|
|
2N98
 
 | | Solution structure of acyl carrier protein LipD from Actinoplanes friuliensis | | Descriptor: | Acyl carrier protein | | Authors: | Paul, S, Ishida, H, Liu, Z, Nguyen, L.T, Vogel, H.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of a freestanding acyl carrier protein involved in the biosynthesis of cyclic lipopeptide antibiotics.
Protein Sci., 26, 2017
|
|
2N99
 
 | | Solution structure of the SLURP-2, a secreted isoform of Lynx1 | | Descriptor: | Ly-6/neurotoxin-like protein 1 | | Authors: | Paramonov, A.S, Shenkarev, Z.O, Lyukmanova, E.N, Arseniev, A.S. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Secreted Isoform of Human Lynx1 (SLURP-2): Spatial Structure and Pharmacology of Interactions with Different Types of Acetylcholine Receptors.
Sci Rep, 6, 2016
|
|
2N9A
 
 | |
2N9B
 
 | |
2N9C
 
 | | NRAS Isoform 5 | | Descriptor: | GTPase NRas | | Authors: | Markowitz, J, Mal, T.K, Yuan, C, Courtney, N.B, Patel, M, Stiff, A.R, Blachly, J, Walker, C, Eisfeld, A, de la Chapelle, A, Carson III, W.E. | | Deposit date: | 2015-11-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of NRAS isoform 5.
Protein Sci., 25, 2016
|
|
2N9D
 
 | |
2N9E
 
 | |
2N9F
 
 | | Glucose as non natural nucleobase | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*(4JA)P*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2N9G
 
 | |
2N9H
 
 | | Glucose as a nuclease mimic in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2N9I
 
 | | Solution structure of reduced human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2N9J
 
 | | Solution structure of oxidized human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9M
 
 | |
2N9N
 
 | |
2N9O
 
 | | Solution structure of RNF126 N-terminal zinc finger domain | | Descriptor: | E3 ubiquitin-protein ligase RNF126, ZINC ION | | Authors: | Martinez-Lumbreras, S, Krysztofinska, E.M, Thapaliya, A, Isaacson, R.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the E3 ligase, RNF126.
Sci Rep, 6, 2016
|
|
