1XC5
 
 | | Solution Structure of the SMRT Deacetylase Activation Domain | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Codina, A, Love, J.D, Li, Y, Lazar, M.A, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2004-09-01 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XCE
 
 | |
1XCY
 
 | | Structure of DNA containing the alpha-anomer of a carbocyclic abasic site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(DXD)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | de los Santos, C, El-khateeb, M, Rege, P, Tian, K, Johnson, F. | | Deposit date: | 2004-09-03 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Impact of the C1 configuration of abasic sites on DNA duplex structure
Biochemistry, 43, 2004
|
|
1XCZ
 
 | | Structure of DNA containing the beta-anomer of a carbocyclic abasic site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(DXD)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | de los Santos, C, El-khateeb, M, Rege, P, Tian, K, Johnson, F. | | Deposit date: | 2004-09-03 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Impact of the C1 configuration of abasic sites on DNA duplex structure
Biochemistry, 43, 2004
|
|
1XDX
 
 | |
1XF7
 
 | | High Resolution NMR Structure of the Wilms' Tumor Suppressor Protein (WT1) Finger 3 | | Descriptor: | Wilms' Tumor Protein, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Dong, J, Huang, K, Carey, P, Weiss, M.A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Why zinc fingers prefer zinc: ligand-field symmetry and the hidden thermodynamics of metal ion selectivity
Biochemistry, 43, 2004
|
|
1XFE
 
 | | Solution structure of the LA7-EGFA pair from the LDL receptor | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor | | Authors: | Beglova, N, Jeon, H, Fisher, C, Blacklow, S.C. | | Deposit date: | 2004-09-14 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Cooperation between Fixed and Low pH-Inducible Interfaces Controls Lipoprotein Release by the LDL Receptor
Mol.Cell, 16, 2004
|
|
1XFL
 
 | | Solution Structure of Thioredoxin h1 from Arabidopsis Thaliana | | Descriptor: | Thioredoxin h1 | | Authors: | Peterson, F.C, Lytle, B.L, Sampath, S, Vinarov, D, Tyler, E, Shahan, M, Markley, J.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-15 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thioredoxin h1 from Arabidopsis thaliana.
Protein Sci., 14, 2005
|
|
1XFN
 
 | | NMR structure of the ground state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1XFQ
 
 | | structure of the blue shifted intermediate state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1XFR
 
 | |
1XGA
 
 | |
1XGB
 
 | |
1XGC
 
 | |
1XGL
 
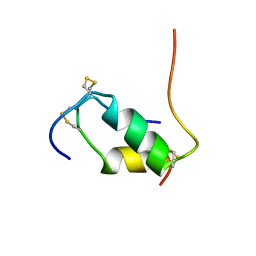 | | HUMAN INSULIN DISULFIDE ISOMER, NMR, 10 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Hua, Q.X, Gozani, S.N, Chance, R.E, Hoffmann, J.A, Frank, B.H, Weiss, M.A. | | Deposit date: | 1996-10-10 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of a protein in a kinetic trap.
Nat.Struct.Biol., 2, 1995
|
|
1XHH
 
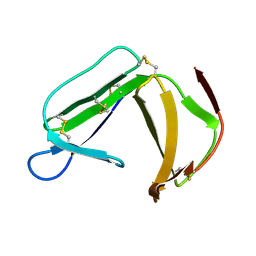 | | Solution Structure of porcine beta-microseminoprotein | | Descriptor: | beta-microseminoprotein | | Authors: | Wang, I, Lou, Y.C, Wu, K.P, Wu, S.H, Chang, W.C, Chen, C. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Novel solution structure of porcine beta-microseminoprotein
J.Mol.Biol., 346, 2005
|
|
1XHJ
 
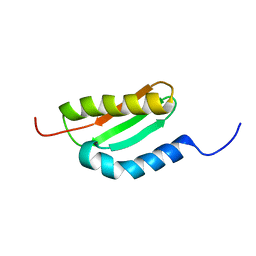 | | Solution Structure Of The Staphylococcus Epidermidis Protein SE0630. Northest Structural Genomics Consortium Target SeR8. | | Descriptor: | Nitrogen Fixation Protein NifU | | Authors: | Baran, M.C, Huang, Y.P, Acton, T, Xiao, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of The Staphylococcus Epidermidis Protein SE0630.
Northest Strucutral Genomics Consortium Target SeR8.
To be Published
|
|
1XHP
 
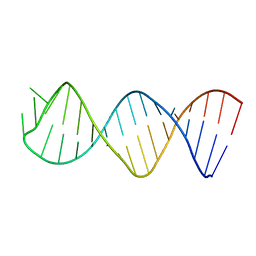 | |
1XHS
 
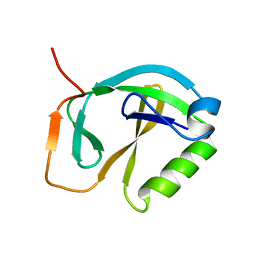 | | Solution NMR Structure of Protein ytfP from Escherichia coli. Northeast Structural Genomics Consortium Target ER111. | | Descriptor: | Hypothetical UPF0131 protein ytfP | | Authors: | Aramini, J.M, Huang, Y.J, Swapna, G.V.T, Paranji, R.K, Xiao, R, Shastry, R, Acton, T.B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Escherichia coli ytfP expands the structural coverage of the UPF0131 protein domain family.
Proteins, 68, 2007
|
|
1XJ1
 
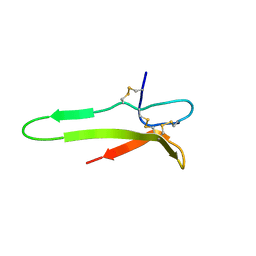 | | 3D solution structure of the C-terminal cysteine-rich domain of the VHv1.1 polydnaviral gene product | | Descriptor: | cysteine-rich omega-conotoxin homolog VHv1.1 | | Authors: | Einerwold, J, Jaseja, J, Hapner, K, Webb, B, Copie, V. | | Deposit date: | 2004-09-22 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carboxyl-terminal cysteine-rich domain of the VHv1.1 polydnaviral gene product: comparison with other cystine knot structural folds
Biochemistry, 40, 2001
|
|
1XJH
 
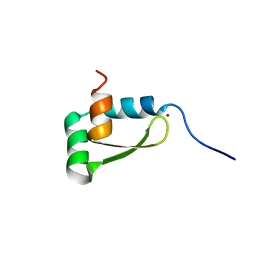 | | NMR structure of the redox switch domain of the E. coli Hsp33 | | Descriptor: | 33 kDa chaperonin, ZINC ION | | Authors: | Won, H.S, Low, L.Y, De Guzman, R.N, Martinez-Yamout, M.A, Jakob, U, Dyson, H.J. | | Deposit date: | 2004-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Zinc-dependent Redox Switch Domain of the Chaperone Hsp33 has a Novel Fold
J.Mol.Biol., 341, 2004
|
|
1XJS
 
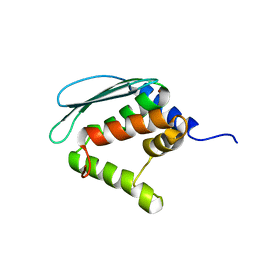 | | Solution structure of Iron-Sulfur cluster assembly protein IscU from Bacillus subtilis, with Zinc bound at the active site. Northeast Structural Genomics Consortium Target SR17 | | Descriptor: | NifU-like protein, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Iron-Sulfur cluster assembly protein IscU
from Bacillus subtilis, with Zinc bound at the active site.
Northeast Structural Genomics Consortium Target SR17.
To be Published
|
|
1XKM
 
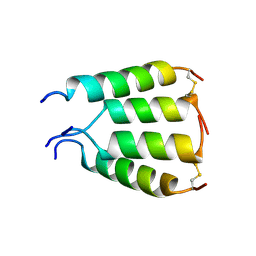 | | NMR structure of antimicrobial peptide distinctin in water | | Descriptor: | Distinctin chain A, Distinctin chain B | | Authors: | Amodeo, P, Raimondo, D, Andreotti, G, Motta, A, Scaloni, A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A folding-dependent mechanism of antimicrobial peptide resistance to degradation unveiled by solution structure of distinctin.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XMW
 
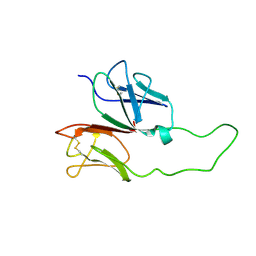 | | CD3 EPSILON AND DELTA ECTODOMAIN FRAGMENT COMPLEX IN SINGLE-CHAIN CONSTRUCT | | Descriptor: | Chimeric CD3 mouse Epsilon and sheep Delta Ectodomain Fragment Complex | | Authors: | Sun, Z.-Y.J, Kim, S.T, Kim, I.C, Fahmy, A, Reinherz, E.L, Wagner, G. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1XN5
 
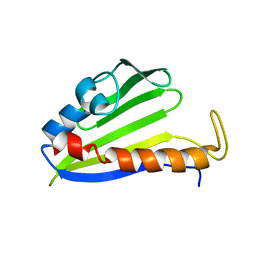 | | Solution Structure of Bacillus halodurans Protein BH1534: The Northeast Structural Genomics Consortium Target BhR29 | | Descriptor: | BH1534 unknown conserved protein | | Authors: | Liu, G, Ma, L, Shen, Y, Acton, T, Atreya, H.S, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Bacillus halodurans Protein BH1534: The Northeast Structural Genomics Consortium Target BhR29
To be Published
|
|
