1S75
 
 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
1S7E
 
 | | Solution structure of HNF-6 | | Descriptor: | Hepatocyte nuclear factor 6 | | Authors: | Liao, X, Sheng, W. | | Deposit date: | 2004-01-29 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the hepatocyte nuclear factor 6alpha and its interaction with DNA.
J.Biol.Chem., 279, 2004
|
|
1S7P
 
 | | Solution structure of thermolysin digested microcin J25 | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Blond, A, Afonso, C, Tabet, J.C, Rebuffat, S, Craik, D.J. | | Deposit date: | 2004-01-30 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of thermolysin cleaved microcin J25: extreme stability of a two-chain antimicrobial peptide devoid of covalent links
Biochemistry, 43, 2004
|
|
1S8K
 
 | | Solution Structure of BmKK4, A Novel Potassium Channel Blocker from Scorpion Buthus martensii Karsch, 25 structures | | Descriptor: | Toxin BmKK4 | | Authors: | Zhang, N, Chen, X, Li, M, Cao, C, Wang, Y, Hu, G, Wu, H. | | Deposit date: | 2004-02-02 | | Release date: | 2005-02-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK4, the first member of subfamily alpha-KTx 17 of scorpion toxins
Biochemistry, 43, 2004
|
|
1S9L
 
 | | NMR Solution Structure of a Parallel LNA Quadruplex | | Descriptor: | 5'-((TLN)P*(LCG)P*(LCG)P*(LCG)P*(TLN))-3' | | Authors: | Randazzo, A, Esposito, V, Ohlenschlager, O, Ramachandran, R, Mayola, L. | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a parallel LNA quadruplex.
Nucleic Acids Res., 32, 2004
|
|
1S9S
 
 | |
1SA8
 
 | |
1SAA
 
 | | ATF-2 RECOGNITION SITE, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*TP*GP*AP*CP*GP*TP*CP*AP*CP*AP*TP*G)-3') | | Authors: | Conte, M.R, Lane, A.N, Bloomberg, G. | | Deposit date: | 1997-08-19 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ATF-2 recognition site and its interaction with the ATF-2 peptide.
Nucleic Acids Res., 25, 1997
|
|
1SAF
 
 | |
1SB0
 
 | | Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb | | Descriptor: | protein CBP, protein c-Myb | | Authors: | Zor, T, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the KIX Domain of CBP Bound to the Transactivation
Domain of c-Myb
J.Mol.Biol., 337, 2004
|
|
1SB6
 
 | | Solution structure of a cyanobacterial copper metallochaperone, ScAtx1 | | Descriptor: | copper chaperone ScAtx1 | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.C, Borrelly, G.P, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-02-10 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a Cyanobacterial Metallochaperone: INSIGHT INTO AN ATYPICAL COPPER-BINDING MOTIF.
J.Biol.Chem., 279, 2004
|
|
1SBJ
 
 | |
1SBU
 
 | | NMR structure of a peptide containing a dimetylthiazolidine : an analog of delta conotoxin EVIA loop 2 | | Descriptor: | delta-conotoxin EVIA | | Authors: | Figuet, M, Chierici, S, Jourdan, M, Dumy, P. | | Deposit date: | 2004-02-11 | | Release date: | 2004-02-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A case study of 2,2-dimethylthiazolidine as locked cis proline amide bond: synthesis, NMR and molecular modeling studies of a [small delta]-conotoxin EVIA peptide analog.
Org.Biomol.Chem., 2, 2004
|
|
1SCV
 
 | |
1SDF
 
 | | SOLUTION STRUCTURE OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STROMAL CELL-DERIVED FACTOR-1 | | Authors: | Crump, M.P, Rajarathnam, K, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
EMBO J., 16, 1997
|
|
1SE7
 
 | | Solution structure of the E. coli bacteriophage P1 encoded HOT protein: a homologue of the theta subunit of E. coli DNA polymerase III | | Descriptor: | HOMOLOGUE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Chikova, A.K, Schaaper, R.M, London, R.E. | | Deposit date: | 2004-02-16 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Phage Like It HOT: Solution Structure of the Bacteriophage P1-Encoded HOT Protein, a Homolog of the theta Subunit of E. coli DNA Polymerase III
Structure, 12, 2004
|
|
1SE9
 
 | |
1SF0
 
 | |
1SF1
 
 | |
1SFV
 
 | | PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B, Tessari, M, Boelens, R, Dijkman, R, Kaptein, R, De Haas, G.H, Verheij, H.M. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2 complexed with micelles and a competitive inhibitor.
J.Biomol.NMR, 5, 1995
|
|
1SG7
 
 | | NMR solution structure of the putative cation transport regulator ChaB | | Descriptor: | Putative Cation transport regulator chaB | | Authors: | Osborne, M.J, Siddiqui, N, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ChaB, a putative membrane ion antiporter regulator from Escherichia coli
BMC STRUCT.BIOL., 4, 2004
|
|
1SGO
 
 | | NMR Structure of the human C14orf129 gene product, HSPC210. Northeast Structural Genomics target HR969. | | Descriptor: | Protein C14orf129 | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shih, L.-Y, Ma, L.-C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the human C14orf129 gene product, HSPC210. Northeast Structural Genomics target HR969.
To be Published
|
|
1SH1
 
 | |
1SHI
 
 | |
1SIY
 
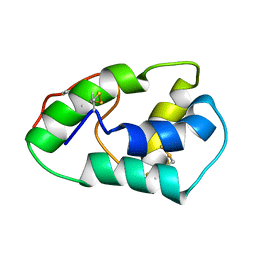 | | NMR structure of mung bean non-specific lipid transfer protein 1 | | Descriptor: | Nonspecific lipid-transfer protein 1 | | Authors: | Lin, K.F, Liu, Y.N, Hsu, S.T.D, Samuel, D, Cheng, C.S, Bonvin, A.M.J.J, Lyu, P.C. | | Deposit date: | 2004-03-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Characterization and Structural Analyses of Nonspecific Lipid Transfer Protein 1 from Mung Bean
Biochemistry, 44, 2005
|
|
