1ATO
 
 | | THE STRUCTURE OF THE ISOLATED, CENTRAL HAIRPIN OF THE HDV ANTIGENOMIC RIBOZYME, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*CP*CP*UP*CP*CP*UP*CP*GP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Kolk, M.H, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1997-08-14 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the isolated, central hairpin of the HDV antigenomic ribozyme: novel structural features and similarity of the loop in the ribozyme and free in solution.
EMBO J., 16, 1997
|
|
1ATV
 
 | | HAIRPIN WITH AGAA TETRALOOP, NMR, 4 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*AP*CP*CP*AP*GP*AP*AP*GP*GP*UP*CP*CP*CP*G)-3') | | Authors: | Kang, H. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Primary Sequence at the Junction of Stem and Loop in RNA Hairpins Affects the Three-Dimensional Conformation in Solution
To be Published
|
|
1ATW
 
 | | HAIRPIN WITH AGAU TETRALOOP, NMR, 3 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*CP*UP*CP*CP*AP*GP*AP*UP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Kang, H. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Primary Sequence at the Junction of Stem and Loop in RNA Hairpins Affects the Three-Dimensional Conformation in Solution
To be Published
|
|
1ATX
 
 | |
1AU5
 
 | | SOLUTION STRUCTURE OF INTRASTRAND CISPLATIN-CROSSLINKED DNA OCTAMER D(CCTG*G*TCC):D(GGACCAGG), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*TP*GP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*CP*AP*GP*G)-3') | | Authors: | Yang, D, Van Boom, S.S.G.E, Reedijk, J, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1997-09-11 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structure and isomerization of an intrastrand cisplatin-cross-linked octamer DNA duplex by NMR analysis.
Biochemistry, 34, 1995
|
|
1AU6
 
 | | SOLUTION STRUCTURE OF DNA D(CATGCATG) INTERSTRAND-CROSSLINKED BY BISPLATIN COMPOUND (1,1/T,T), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIS(TRANS-PLATINUM ETHYLENEDIAMINE DIAMINE CHLORO)COMPLEX, DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3') | | Authors: | Yang, D, Van Boom, S.S.G.E, Reedijk, J, Van Boom, J.H, Farrell, N, Wang, A.H.-J. | | Deposit date: | 1997-09-11 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel DNA structure induced by the anticancer bisplatinum compound crosslinked to a GpC site in DNA.
Nat.Struct.Biol., 2, 1995
|
|
1AUD
 
 | | U1A-UTRRNA, NMR, 31 STRUCTURES | | Descriptor: | RNA 3UTR, U1A 102 | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1997-08-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the RNA-binding specificity of human U1A protein.
EMBO J., 16, 1997
|
|
1AUZ
 
 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AV3
 
 | | POTASSIUM CHANNEL BLOCKER KAPPA CONOTOXIN PVIIA FROM C. PURPURASCENS, NMR, 20 STRUCTURES | | Descriptor: | Kappa-conotoxin PVIIA | | Authors: | Scanlon, M.J, Naranjo, D, Thomas, L, Alewood, P.F, Lewis, R.J, Craik, D.J. | | Deposit date: | 1997-09-24 | | Release date: | 1998-10-14 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Solution structure and proposed binding mechanism of a novel potassium channel toxin kappa-conotoxin PVIIA.
Structure, 5, 1997
|
|
1AW6
 
 | | GAL4 (CD), NMR, 24 STRUCTURES | | Descriptor: | CADMIUM ION, GAL4 (CD) | | Authors: | Baleja, J.D, Wagner, G. | | Deposit date: | 1997-10-10 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the DNA-binding domain of GAL4 and use of 3J(113Cd,1H) in structure determination.
J.Biomol.NMR, 10, 1997
|
|
1AWW
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, 42 STRUCTURES | | Descriptor: | BRUTON'S TYROSINE KINASE | | Authors: | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
1AWX
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BRUTON'S TYROSINE KINASE | | Authors: | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
1AWY
 
 | | NMR STRUCTURE OF CALCIUM BOUND CONFORMER OF CONANTOKIN G, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CONANTOXIN G | | Authors: | Rigby, A.C, Baleja, J.D, Leping, L, Pedersen, L.G, Furie, B.C, Furie, B. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Role of gamma-carboxyglutamic acid in the calcium-induced structural transition of conantokin G, a conotoxin from the marine snail Conus geographus.
Biochemistry, 36, 1997
|
|
1AX6
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT OPPOSITE A-2 DELETION SITE IN THE NARI HOT SPOT SEQUENCE CONTEXT; NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(CTCGGC-[AF]G-CCATC)D(GATGGCCGAG) | | Authors: | Mao, B, Gorin, A.A, Gu, Z, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a--2 deletion site in the NarI hot spot sequence context.
Biochemistry, 36, 1997
|
|
1AX7
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED AT A TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(AAC-[AF]G-CTACCATCC)D(GGATGGTAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-stacked conformer of the syn [AF]-C8-dG adduct positioned at a template-primer junction.
Biochemistry, 36, 1997
|
|
1AXH
 
 | | ATRACOTOXIN-HVI FROM HADRONYCHE VERSUTA (AUSTRALIAN FUNNEL-WEB SPIDER, NMR, 20 STRUCTURES | | Descriptor: | ATRACOTOXIN-HVI | | Authors: | Fletcher, J.I, O'Donoghue, S.I, Nilges, M, King, G.F. | | Deposit date: | 1996-11-04 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structure of a novel insecticidal neurotoxin, omega-atracotoxin-HV1, from the venom of an Australian funnel web spider.
Nat.Struct.Biol., 4, 1997
|
|
1AXJ
 
 | |
1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | Authors: | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
1AXO
 
 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
1AXP
 
 | | DNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 6 STRUCTURES | | Descriptor: | DNA (D(GAAGAGAAGC)(DOT)D(GCTTCTCTTC)) | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-17 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1AXU
 
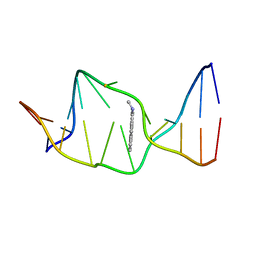 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AY3
 
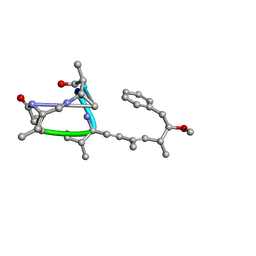 | | Nodularin from Nodularia spumigena | | Descriptor: | PEPTIDIC TOXIN NODULARIN | | Authors: | Annila, A.J. | | Deposit date: | 1997-11-14 | | Release date: | 1999-08-16 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of nodularin. An inhibitor of serine/threonine-specific protein phosphatases.
J.Biol.Chem., 271, 1996
|
|
1AYJ
 
 | |
1AYK
 
 | |
