9CPI
 
 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[1-(2-aminoethyl)-4,5-dihydro-1H-imidazol-2-yl]-N-[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]benzamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
9CSG
 
 | | Human Serum Albumin Bound to Cerastecin Compound 5e | | Descriptor: | 2-(4-butylbenzene-1-sulfonamido)-5-(4-{3-carboxy-4-[4-(2-methoxyethyl)benzene-1-sulfonamido]anilino}-4-oxobutanamido)benzoic acid, Albumin, MYRISTIC ACID | | Authors: | Hruza, A, Klein, D.J, Ishchenko, A. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
9CSI
 
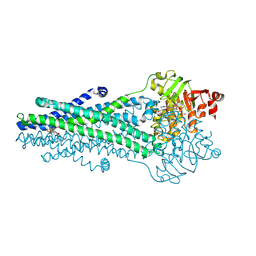 | | A. baumannii MsbA Bound to Cerastecin Compound 5 | | Descriptor: | 3,3'-[(1,4-dioxobutane-1,4-diyl)bis(azanediyl)]bis[(4-butylbenzene-1-sulfonamido)benzoic acid], Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Klein, D.J, Ishchenko, A, Soisson, S, Cheng, R, Hennig, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
9CSY
 
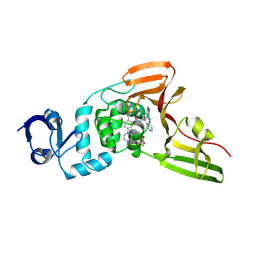 | | SARS-CoV-2 papain-like protease (PLpro) bound to PF-07957472 | | Descriptor: | 2-methyl-5-(4-methylpiperazin-1-yl)-N-{1-[(2P)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]cyclopropyl}benzamide, Papain-like protease, ZINC ION, ... | | Authors: | Mashalidis, E.H, Chang, J.S, Wu, H, Garnsey, M. | | Deposit date: | 2024-07-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Discovery of SARS-CoV-2 papain-like protease (PL pro ) inhibitors with efficacy in a murine infection model.
Sci Adv, 10, 2024
|
|
9CTH
 
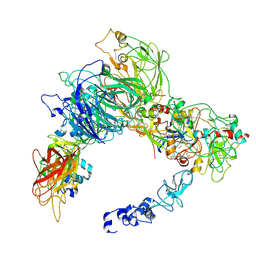 | | Preliminary map of the Prothrombin-prothrombinase complex on nano discs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activated Factor V (FVa) heavy chain, Activated Factor V (FVa) light chain, ... | | Authors: | Stojanovski, B.M, Mohammed, B.M, Di Cera, E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | The Prothrombin-Prothrombinase Interaction.
Subcell Biochem, 104, 2024
|
|
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 2024
|
|
9CUW
 
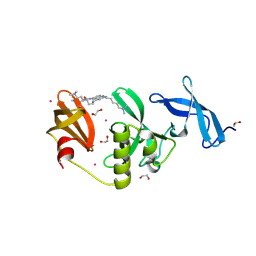 | | Crystal Structure of SETDB1 Tudor domain in complex with UNC100013 | | Descriptor: | (2E)-4-(dimethylamino)-N-(4-{[6-(dimethylamino)hexyl]amino}-2-{[5-(dimethylamino)pentyl]amino}quinazolin-6-yl)but-2-enamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Silva, M, Dong, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-26 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of SETDB1 Tudor domain in complex with UNC100013
To be published
|
|
9CUX
 
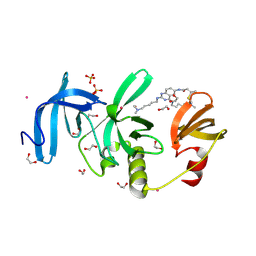 | | Crystal Structure of SETDB1 Tudor domain in complex with UNC100016 | | Descriptor: | (2E)-N-(4-{[6-(dimethylamino)hexyl]amino}-2-{[5-(dimethylamino)pentyl]amino}quinazolin-6-yl)but-2-enamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Silva, M, Dong, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-26 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal Structure of SETDB1 Tudor domain in complex with UNC100016
To be published
|
|
9CUZ
 
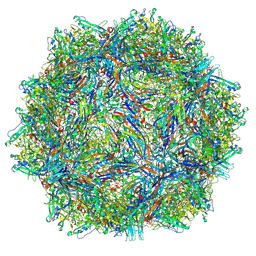 | | Bufavirus 1 complexed with 6SLN | | Descriptor: | N-acetyl-alpha-neuraminic acid, VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-27 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9CV0
 
 | | Bufavirus 1 at pH 7.4 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-27 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9CV9
 
 | | Bufavirus 1 at pH 4.0 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-28 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9CWS
 
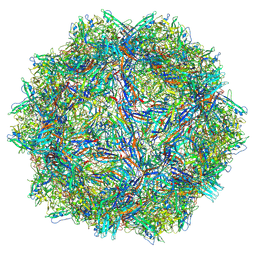 | | Bufavirus 1 at pH 2.6 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9D3E
 
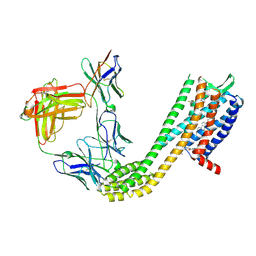 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM2 | | Descriptor: | 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CHOLESTEROL, Human CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
9D3G
 
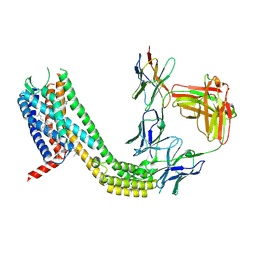 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM1 | | Descriptor: | 1-(4-chlorophenyl)-N-{[(2R)-4-(2,3-dihydro-1H-inden-2-yl)-5-oxomorpholin-2-yl]methyl}cyclopropane-1-carboxamide, 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
9D5D
 
 | |
9D77
 
 | | Crystal form of Netrin-1 mimics nanotubes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Meier, M, Krahn, N.J, McDougall, M.D, Rafiei, F, Koch, M, Stetefeld, J. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanistic insights in the higher-order protein assemblies of netrin-1
To Be Published
|
|
9D93
 
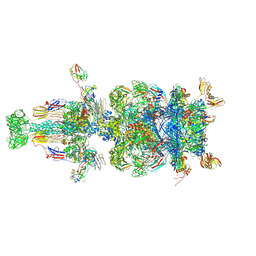 | |
9D94
 
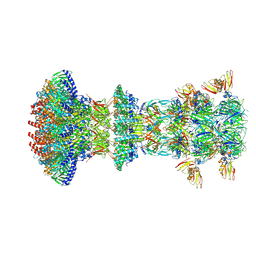 | |
9D9K
 
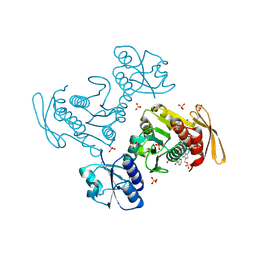 | |
9D9W
 
 | |
9D9X
 
 | |
9DHG
 
 | | DHODH in complex with Compound 5 | | Descriptor: | (3P,7P)-7-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-6-fluoro-3-(2-methylphenyl)-1-(propan-2-yl)quinolin-4(1H)-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Identification of isoquinolinone DHODH inhibitor isosteres.
Bioorg.Med.Chem.Lett., 2024
|
|
9DHH
 
 | | DHODH in complex with Compound 8 | | Descriptor: | (3S,7P)-7-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-6-fluoro-3-(2-methylphenyl)-1-(propan-2-yl)-2,3-dihydroquinolin-4(1H)-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification of isoquinolinone DHODH inhibitor isosteres.
Bioorg.Med.Chem.Lett., 2024
|
|
9DHI
 
 | | DHODH in complex with Compound 7 | | Descriptor: | (2M,6P)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-2-(2-methylphenyl)-4-(propan-2-yl)phthalazin-1(2H)-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification of isoquinolinone DHODH inhibitor isosteres.
Bioorg.Med.Chem.Lett., 2024
|
|
9DHJ
 
 | | DHODH in complex with Compound 15 | | Descriptor: | (2M,6P)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-2-(2-methylphenyl)-4-[(2R)-1,1,1-trifluoropropan-2-yl]phthalazin-1(2H)-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of isoquinolinone DHODH inhibitor isosteres.
Bioorg.Med.Chem.Lett., 2024
|
|






